Skip to comments.
Mitochondrial DNA Mutation Rates
University of North Carlolina Computer Science Department website ^
| from 1997 to present
| David Plaistid
Posted on 09/01/2002 4:20:09 PM PDT by Ahban
Mitochondrial DNA Mutation Rates
David A. Plaisted
Recently an attempt was made to estimate the age of the human race using mitochondrial DNA. This material is inherited always from mother to children only. By measuring the difference in mitochondrial DNA among many individuals, the age of the common maternal ancestor of humanity was estimated at about 200,000 years. A problem is that rates of mutation are not known by direct measurement, and are often computed based on assumed evolutionary time scales. Thus all of these age estimates could be greatly in error. In fact, many different rates of mutation are quoted by different biologists.
It shouldn't be very hard explicitly to measure the rate of mutation of mitochondrial DNA to get a better estimate on this age. From royal lineages, for example, one could find two individuals whose most recent common maternal ancestor was, say, 1000 years ago. One could then measure the differences in the mitochondrial DNA of these individuals to bound its mutation rate. This scheme is attractive because it does not depend on radiometric dating or other assumptions about evolution or mutation rates. It is possible that in 1000 years there would be too little difference to measure. At least this would still give us some useful information.
(A project for creation scientists!)
Along this line, some work has recently been done to measure explictly the rate of substitution in mitochondrial DNA. The reference is Parsons, Thomas J., et al., A high observed substitution rate in the human mitochondrial DNA control region, Nature Genetics vol. 15, April 1997, pp. 363-367. The summary follows:
"The rate and pattern of sequence substitutions in the mitochondrial DNA (mtDNA) control region (CR) is of central importance to studies of human evolution and to forensic identity testing. Here, we report a direct measurement of the intergenerational substitution rate in the human CR. We compared DNA sequences of two CR hypervariable segments from close maternal relatives, from 134 independent mtDNA lineages spanning 327 generational events. Ten subsitutions were observed, resulting in an empirical rate of 1/33 generations, or 2.5/site/Myr. This is roughly twenty-fold higher than estimates derived from phylogenetic analyses. This disparity cannot be accounted for simply by substitutions at mutational hot spots, suggesting additional factors that produce the discrepancy between very near-term and long-term apparent rates of sequence divergence. The data also indicate that extremely rapid segregation of CR sequence variants between generations is common in humans, with a very small mtDNA bottleneck. These results have implications for forensic applications and studies of human evolution." (op. cit. p. 363).
The article also contains this section: "The observed substitution rate reported here is very high compared to rates inferred from evolutionary studies. A wide range of CR substitution rates have been derived from phylogenetic studies, spanning roughly 0.025-0.26/site/Myr, including confidence intervals. A study yielding one of the faster estimates gave the substitution rate of the CR hypervariable regions as 0.118 +- 0.031/site/Myr. Assuming a generation time of 20 years, this corresponds to ~1/600 generations and an age for the mtDNA MRCA of 133,000 y.a. Thus, our observation of the substitution rate, 2.5/site/Myr, is roughly 20-fold higher than would be predicted from phylogenetic analyses. Using our empirical rate to calibrate the mtDNA molecular clock would result in an age of the mtDNA MRCA of only ~6,500 y.a., clearly incompatible with the known age of modern humans. Even acknowledging that the MRCA of mtDNA may be younger than the MRCA of modern humans, it remains implausible to explain the known geographic distribution of mtDNA sequence variation by human migration that occurred only in the last ~6,500 years.
One biologist explained the young age estimate by assuming essentially that 19/20 of the mutations in this control region are slightly harmful and eventually will be eliminated from the population. This seems unlikely, because this region tends to vary a lot and therefore probably has little function. In addition, the selective disadvantage of these 19/20 of the mutations would have to be about 1/300 or higher in order to avoid producing more of a divergence in sequences than observed in longer than 6000 years. This means that one in 300 individuals would have to die from having mutations in this region. This seems like a high figure for a region that appears to be largely without function. It is interesting that this same biologist feels that 9/10 of the mutations to coding regions of DNA are neutral. This makes the coding regions of DNA less constrained than the apparently functionless control region of the mitochondrial DNA!
TOPICS: Miscellaneous
KEYWORDS: creationsim; crevolist; dna; evolution; godsgravesglyphs; helixmakemineadouble; history; mtdna; multiregionalism; mutationrates
Navigation: use the links below to view more comments.
first previous 1-20, 21-40, 41-60, 61-80 ... 141-157 next last
To: Ahban
What do you make of this???? A bit dated as it leaves off the now known possibility of male influence on mtDNA. Still, that is a factor that should increase the mutation rate above the accepted level- which is the whole point of this article. You're correct, Ahban. Paternal mtDNA affects would account for the hypermutated regions. The Smith article discusses this as well.
21
posted on
09/01/2002 8:23:24 PM PDT
by
Nebullis
To: Nebullis
bump
To: aruanan
The article in your #12 would indicate that paternal inheiritence of mtDNA is a rare fluke that would not effect the studies cited in my orignial post. This then implies that those mtDNA studies that have "Eve" at 133,000-200,000 years ago are off to the long side by as much as a factor of twenty.
23
posted on
09/02/2002 4:29:38 AM PDT
by
Ahban
To: VadeRetro
I don't think mtDNA can be used to find the evolutinary relationships between, say man and fish - because I am unconvinced any such relationship exists other than code borrowing. I agree with the critques of mtDNA on that (though not for the same reasons they do).
For tracing ancestry of very closely related groups, I think it is a great tool and so do a lot of scientists - that is why they keep using it. Just because it is not a valid tool for finding distant evolutionary relationships (or as I would put it, the technique shows a NEGATIVE RESULT for distant evolutionary relationships) does not mean it is useless for finding out how far in the past our first common ancestor lived.
Looks like it was a lot more recently than 133,000 years ago....
24
posted on
09/02/2002 4:34:54 AM PDT
by
Ahban
To: Verginius Rufus
With all of the things we are accomplishing these days, I would think tracing linaege in this way would be a minor accomplishment, well within our technical ability.
25
posted on
09/02/2002 4:37:45 AM PDT
by
Ahban
To: ThinkPlease
But this HAS been peer reviewed. Not Plaisted's report on it, but his cite,
Nature Genetics vol. 15, April 1997, pp. 363-367. That is peer-reveiwed, right?
So what do you make of this peer-reviewed paper that says molecular clocks are off by a factor of up to 20 on the long side? How much more recently than 133,000 years ago did the common female ancestor of all humans live?
26
posted on
09/02/2002 4:44:25 AM PDT
by
Ahban
To: gore3000
I agree that mtDNA has been proven no good for detected distant evolutionary relationships- but this is simply because there are no such relationships. So far as I know, mtDNA is still good for finding the relationship among people that actually did descend from one another.
mtDNA seems like it is offering stronger support for our position every day. It shows a NEGATIVE RESULT for distant evolutionary relationships(of course they say it just means the tool is no good for detecting the relationship- I say the tool is fine, it is the relatinoship that is no good). In addition, on those relationships it can measure it shows a VERY recent date for mankind.
Even before this, they had to make the assumed population unrealistically small to come up with date from 133,000-200,000 years ago for Eve. Now their problems are compounded.
27
posted on
09/02/2002 4:53:14 AM PDT
by
Ahban
To: Nebullis
Okay, paternal mtDNA is a rare possibility, but it happens. That should not have effected the experiment cited in my post though (the one that showed mutation rates were 20 times higher than assumed.) That experiment did not trace long enough for the seemingly rare paternal mtDNA to be a factor.
If it is an additional factor, that would drive the mutation rate even higher. That means the assumptions most scientists have been making are wrong. That in turn implies that the studies that showed Eve as having lived about 133,000 years ago are way off. With a higher mutation rate the time needed for the observed mutations goes way down.
So, by the numbers, how long ago did Eve live?????
28
posted on
09/02/2002 4:59:53 AM PDT
by
Ahban
To: Ahban
Plaisted seemed to be saying that we should take two individuals whose relationship can be documented from births and marriages of royal families, and compare that with the scientific evidence. My point is that the records are far too incomplete to take that back 1000 years. You might succeed going back 200 or 300 years or even 500, but beyond that to trace from mother to mother to mother will soon become impossible.
In England the government ordered every parish to keep baptismal records only in the 1530s (and not all of the earlier records survive), and England is probably the country with the best records--in Scotland many parishes have no records before the 1700s surviving, and many of the Irish records were destroyed in 1922. Kings did not only marry women from other royal families. Imagine trying to trace Anne Boleyn's lineage from mother to mother back to 1000.
It might be possible to identify people who share the same maternal ancestor 1000 years back on the basis of the DNA evidence, but I suspect you'd have to test a very large number of people to find such a case, and mutations in the intervening millennium might cause the relationship to escape notice. Iceland might be a good place to look since they've had a pretty stable population over the centuries.
To: Ahban
So, by the numbers, how long ago did Eve live????? Er, let's see...133,000/20*creationist fudge factor...by Jove, exactly 6000 years ago!!!!!
mtDNA is loved for its rapid mutation rates. Now it turns out that sections of the D-Loop which have been used for molecular clocking are hypervariable; they mutate at higher than background mtDNA rates. This means that other sections of the D-Loop should be used for estimates farther out. If a rate shift has occurred over a long time period, rate shifts can be taken into account.
30
posted on
09/02/2002 6:51:02 AM PDT
by
Nebullis
To: Ahban
The article in your #12 would indicate that paternal inheiritence [sic] of mtDNA is a rare fluke that would not effect [sic] the studies cited in my orignial [sic] post. This then implies that those mtDNA studies that have "Eve" at 133,000-200,000 years ago are off to the long side by as much as a factor of twenty. [????]
You miss the point. The study cited (as well as others) indicate that the transmission of mitochondrial DNA in a clonal fashion is not as slam-dunk an idea as once supposed. It demonstrates that the transmission of mtDNA in a clonal fashion is uncertain to an
unknown degree. Because of this, strategies the accuracy of which depend on such a transmission become less than certain. By how much? It's not known, hence the concluding paragraph from my post above:
Nevertheless, even a single validated example of paternal mtDNA transmission suggests that the interpretation of inheritance patterns in other kindreds thought to have mitochondrial disease should not be based on the dogmatic assumption of absolute maternal inheritance of mtDNA. Likewise, the possibility of paternal inheritance of mtDNA should be accommodated in statistical models that analyze sequence variations in mtDNA in different human or primate populations in order to draw inferences about human evolution or migration. The unusual case described by Schwartz and Vissing is more than a mere curiosity. [emphasis added]
31
posted on
09/02/2002 7:13:35 AM PDT
by
aruanan
To: Ahban
I don't think mtDNA can be used to find the evolutinary relationships between, say man and fish - because I am unconvinced any such relationship exists other than code borrowing. Talk about circular logic! You don't think we can find a tree because there is no underlying tree relationship. But it's always a tree. It's not always the same exact tree from every method, but the trees tend to be highly similar. In the evolutionary version, that's because the trees with varying degrees of error reflect an underlying reality, a true branching hierarchy of common descent.
We have evidence for short-term changes in species populations, but the deniers chirp that that's just micro-evolution and nobody disputes that. We also have plenty of evidence for long-term change, but that's some kind of code-borrowing in successive creation events.
Code borrowing? By whom? When? In how many distinct events of "creation?" Is somebody running around doing this all the time? Is this somebody making gravity and electromagnetism work too? This hypothesis is cluttered with poorly explained elements.
To: Ahban
For tracing ancestry of very closely related groups, I think it is a great tool and so do a lot of scientists - that is why they keep using it. Just because it is not a valid tool for finding distant evolutionary relationships (or as I would put it, the technique shows a NEGATIVE RESULT for distant evolutionary relationships) does not mean it is useless for finding out how far in the past our first common ancestor lived. Different molecular clocks are useful over different time ranges, yes. For some reason, cytochrome c and the gene that codes it have a very slow mutation rate and can be used to build what amounts to a tree of all life. The tree you get is about what an evolutionist would expect. Humans and chimpls have the exact same molecule, with a single silent mutation in the gene. The yeast Candida krusei, highly unrelated to humans, has 51 amino acid differences in the molecule. (But human cytochrome will work in a yeast which has had its own cytochrome removed.)
To: VadeRetro
Let's ignore my code-borrowing aside for the nonce. How does YOUR hypothesis of human evolution from non-human primates square with the evidence presented in this post? Any extra errors, like paternal mtDNA, would only make the point of this artilce sharper. The assumed mutation rates, the ones that have a rough date of 133,000 years ago, are far to low.
I say that argues for a very recent mankind. So recent that there was nothing around to evolve from. I will discuss the code borrowing with you another time, as I have discussed it in the past, but I did not mean to change the subject with that aside. What about the evidence that is the essense of the this post?
34
posted on
09/02/2002 8:00:10 AM PDT
by
Ahban
To: Nebullis
OK, but you do agree the implications of the data so far is that the common ancestor of all humans is more recent than 133,000 years ago? I agree more data needs to be collected, but so far what the data seems to be saying is that humans are very recent. You said we need to measure some other loop for long range because this one mutates too fast, but don't we need to be careful not to misjudge the mutation rates of the new loops we look at?
And how long will it go on? I mean, if you keep looking at loops, keep putting in assumed mutation rates that are too low and don't find out about it for a decade........ How long will this go on before scientists just admit mankind is young. Or will they just knock over the chessboard and say mtDNA is no good at all because it does not yield the expected result?
35
posted on
09/02/2002 8:08:36 AM PDT
by
Ahban
To: Ahban
How does YOUR hypothesis of human evolution from non-human primates square with the evidence presented in this post? mtDNA is just one tool. Some guy found one hypervariable site. You're announcing it's all a house of cards. So far, I don't see where that follows. If I understand "hypervariable," the overall mutation rate in mtDNA is still far lower than for this "hypervariable" region. Do most studies focus on this region?
For sure, there's other evidence for humans arising from primates.
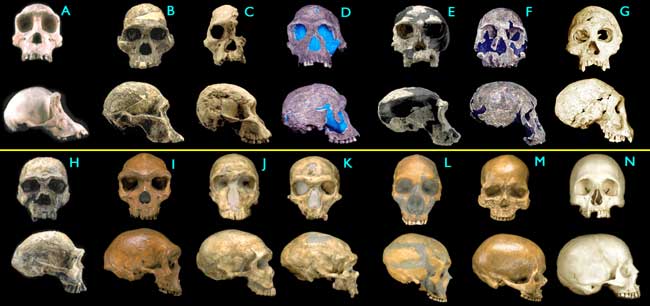
Discussion.
To: aruanan
I don't think I am missing the point at all. I understand that the possibilty of paternal inheiriteince of mtDNA should be taken into account in divergence studies.
Look at at this line from the article.
We compared DNA sequences of two CR hypervariable segments from close maternal relatives, from 134 independent mtDNA lineages spanning 327 generational events
They looked at 134 people for about 3 generations each. If paternal mtDNA, a rare occurance even it does happen, skewed the rates at all, it would have showed up as a pile of mutations in one of the cases. They would have spotted that and thrown it out. My point remains that the mutation rates found in THIS CITE were way above that needed for a 133,000 year old man, and that it is unlikely that paternal mtDNA affected this study.
So I agree with you and your cite that paternal mtDNA may need to be taken into account. I don't think it was a factor in the study cited in my post though....and that is the problem I hope you will grapple with.
37
posted on
09/02/2002 8:22:43 AM PDT
by
Ahban
To: Ahban
OK, but you do agree the implications of the data so far is that the common ancestor of all humans is more recent than 133,000 years ago?No, there's no evidence for that. If a specific region of mtDNA is inaccurate for measurements in a estimated age-range (based on other, confirming dating), or if the error estimates are larger than what you want to measure, then that specific region simply doesn't tell you much of anything. It doesn't mean that actual distances are shorter. It means different sections of DNA or larger data sets need to be used for a better clock.
38
posted on
09/02/2002 8:26:18 AM PDT
by
Nebullis
To: VadeRetro
Yes. It is my understanding that this section of mtDNA was the main one they looked at for short term events because they knew it was a rapid mutator. They just did not know HOW rapid.
I agree that the bones are your strongest point. The bones are not a slam-dunk however. They are far too subjective and inconclusive for that. Any graduated series can look like an evolutinoary progression. THat does not establish that it is. Convergence can mimic direct descent. I just think that the DNA evidnece is far more powerful and less subjective than bones. Especially since many of those are dated by luminessence, which only gives a MAXIMUM age which the press treats as the actual age of the finds.
Bones vs. DNA. Those two are not a tie, and if they are, cultural evidence, art and especially religion, show a 'big bang' in the more recent past. This breaks the tie, but I agree the contest is not over. I am not asking you to say you lost, just admit we are scoring a touchdown now and then.
39
posted on
09/02/2002 8:34:21 AM PDT
by
Ahban
To: Ahban
Convergence can mimic direct descent. Convergence leaves a different fossil record. And even the most spectacular convergences like the Thylacine are more or less instantly detectable.
I'm showing you apes morphing into humans. No distinct border across "created kinds" is apparent anywhere in the sequence. You're giving me "Mumble, mumble."
Navigation: use the links below to view more comments.
first previous 1-20, 21-40, 41-60, 61-80 ... 141-157 next last
Disclaimer:
Opinions posted on Free Republic are those of the individual
posters and do not necessarily represent the opinion of Free Republic or its
management. All materials posted herein are protected by copyright law and the
exemption for fair use of copyrighted works.
FreeRepublic.com is powered by software copyright 2000-2008 John Robinson
