Skip to comments.
Biomimicry: why the world is full of intelligent design (they admit ID, then credit evolution!!!)
Telegraph ^
| June 8, 2009
| Sanjida O'Connell
Posted on 06/08/2009 4:41:47 PM PDT by GodGunsGuts
Biomimicry: why the world is full of intelligent design
Forget human ingenuity - the best source of ideas for cutting-edge technology might be in nature, according to experts in 'biomimicry'
We humans like to think we're pretty good at design and technology – but we often forget that Mother Nature had a head start of 3.6 million years. Now, the way that geckoes climb walls, or hummingbirds hover, is at the centre of a burgeoning industry: biomimicry, the science of "reverse-engineering" clever ideas from the natural world....
(Excerpt) Read more at telegraph.co.uk ...
TOPICS: Business/Economy; Culture/Society; Miscellaneous; News/Current Events
KEYWORDS: catholic; christian; creation; darwindrones; darwiniacs; economy; evolution; evoplagiarism; evoreligion; evotheft; fools; godsgravesglyphs; goodgodimnutz; intelligentdesign; naturalselectiongod; oldtimeevoreligion; romans120; science; templeofdarwin
Navigation: use the links below to view more comments.
first previous 1-20 ... 141-160, 161-180, 181-200 ... 261-271 next last
To: Fichori
How do you propose that a 2% genetic change between a human and a chimp would be “constrained”?
Where is the barrier to survivability in changing our DNA by that amount? What is the constraint?
And these “critters” were designed to EVOLVE through mutation, not “adapt” through mutations that are somehow constrained.
Adaptation is a change in how DNA is used (expressing more melanin in response to laying out in the Sun), evolution is a change in the DNA itself.
161
posted on
06/12/2009 3:11:13 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: Dutchboy88
What exactly do you think evolution is if not random mutation, natural selection and common descent? All of these rely upon no design, direction, control or management. So, what part of this would a Designer implement?In three sentences you have managed to convince the world that you are totally ignorant on the theory of evolution.
To: AndrewC
A molecule hits a receptor that activates a transcription factor that turns on a gene.
That is a signal transduction pathway. Nowhere along that pathway was there a choice or an intelligent decision by the cell.
If you don't think mutational hotspots are hotspots because they are difficult to replicate with 100% fidelity, how do YOU explain what makes them “hot” for mutation?
As I said, some sequences are just more likely to get mutated than others, and the reason is, like a tongue twister, some combinations are difficult to do sequentially.
163
posted on
06/12/2009 3:14:44 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: AndrewC
52 ! = 8.06581752 × 1067I haven't checked your math but that would be the same odds for any sequence. The same as for the sequence that I just dealt. It happened even though the odds of it happening are 52 ! = 8.06581752 × 106752 ! = 8.06581752 × 1067 according to you!
You are busted.
To: allmendream
“How do you propose that a 2% genetic change between a human and a chimp …” [excerpt]
Not only has your 2% been
shown to be a dishonest card trick, you are also changing the subject.
“Where is the barrier to survivability in changing our DNA by that amount?” [excerpt]
Your question is based on your flawed 2%.
“And these “critters” were designed to EVOLVE through mutation, ” [excerpt]
Statement of faith.
Nothing more, nothing less.
“Adaptation is …” [excerpt]
Adaptation is whatever you need it to be.
165
posted on
06/12/2009 3:44:58 PM PDT
by
Fichori
To: Fichori
It is not a “dishonest card trick” to take the DNA sequence of a gene in chimps and the DNA sequence for the “same” gene in humans and compare them letter by letter.
When you compare a human gene to the homologous gene in chimps it is about 98% the same. Go to PubMed and blast a gene sequence if you don't want to take my word for it. Do you want to “go on record” as saying the answer will be anything other than about 98%?
How is a chimpanzee genetic DNA “constrained” against a 2% change into a different, yet still perfectly functional, gene?
If DNA is being changed within the population that is evolution.
If how the DNA is being adjusted to fit circumstances that is usually referred to as adaptation.
If you admit that DNA is being changed in the population in order to “adapt” to circumstances (selective pressure), that is evolution.
My entire point in talking about error prone DNA polymerase has been to illustrate the utility of generating genetic diversity in order to change the DNA of a population such that it evolves to fit circumstances.
That is evolution.
166
posted on
06/12/2009 3:59:25 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: ColdWater
167
posted on
06/12/2009 4:20:41 PM PDT
by
AndrewC
(Metanoia)
To: allmendream; CottShop
“It is not a “dishonest card trick” to take the DNA sequence of a gene in chimps and the DNA sequence for the “same” gene in humans and compare them letter by letter.” [excerpt]
From the
link I posted earlier:
The availability of the chimp genome sequence in 2005 has provided a more realistic comparison. It should be noted that the chimp genome was sequenced to a much less stringent level than the human genome, and when completed it initially consisted of a large set of small un-oriented and random fragments. To assemble these DNA fragments into contiguous sections that represented large regions of chromosomes, the human genome was used as a guide or framework to anchor and orient the chimp sequence. Thus, the evolutionary assumption of a supposed ape to human transition was used to assemble the otherwise random chimp genome.
Comparing chimp data that has been arranged to look like human data, to human data is a dishonest card trick.
“When you compare a human gene to the homologous gene in chimps it is about 98% the same.” [excerpt]
Have they resequenced the chimp genome
objectively?
Until they do, its just fudged data.
“If how the DNA is being adjusted to fit circumstances that is usually referred to as adaptation.” [excerpt]
Well, I guess the people who sequenced the chimp genome were just
adapting it to fit their circumstances.
“If you admit that DNA is being changed in the population in order to “adapt” to circumstances (selective pressure), that is evolution.” [excerpt]
Oh, its being changed all right … by Evolutionists after the fact.
And yes, Evolution
is predominantly about
adapting and
changing the facts.
168
posted on
06/12/2009 4:21:35 PM PDT
by
Fichori
To: Fichori
Once again you are talking about GENOMIC comparisons, which has estimated to be abut 90-96% the same.
I am talking directly sequenced GENES. Genetic DNA is about 98% the same between humans and chimps.
http://news.nationalgeographic.com/news/2005/08/0831_050831_chimp_genes_2.html
“Eichler and his colleagues found that the human and chimp sequences differ by only 1.2 percent in terms of single-nucleotide changes to the genetic code.”
“our closest living relatives share 96 percent of our DNA. The number of genetic differences between humans and chimps is ten times smaller than that between mice and rats.”
So back to the subject of your supposed constraint. What would you imagine would be the constraint on a mouse's DNA that would prevent it evolving into something like a rat?
169
posted on
06/12/2009 4:57:34 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: allmendream
A molecule hits a receptor that activates a transcription factor that turns on a gene.It is a bit more complex then you make it out to be. What is the difference between a repression and de-repression?
From the previous article.
A central part of the SOS response is the de-repression of more than 20 genes under the direct and indirect transcriptional control of the LexA repressor. The LexA regulon includes recombination and repair genes recA, recN, and ruvAB, nucleotide excision repair genes uvrAB and uvrD, the error-prone DNA polymerase (pol) genes dinB (encoding pol IV) (2) and umuDC (encoding pol V) (3), and DNA polymerase II (4, 5) in addition to many functions not yet understood. In the absence of a functional SOS response, cells are sensitive to DNA damaging agents.
The signal transduction pathway leading to an SOS response (reviewed by ref. 6) ensues when RecA protein binds to single-stranded DNA (ssDNA), which can be created by processing of DNA damage, stalled replication, and perhaps by other means (7–9). The ssDNA acts as a signal that activates an otherwise dormant co-protease activity of RecA, which allows activated RecA (called RecA*) to facilitate the proteolytic self-cleavage of the LexA repressor, thus inducing the LexA regulon (10). Activated RecA also facilitates the cleavage of phage repressors used to maintain the quiescent, lysogenic state, and UmuD, creating UmuD′, the subunit of UmuD′C (pol V) that allows activity in trans-lesion error-prone DNA synthesis (6).
...
We have examined the role of the SOS response in adaptive mutation and report both positive and negative control of adaptive mutation in the Lac system by the LexA repressor. First, we report that SOS induction of the LexA regulon is required for efficient adaptive mutation. Simple overproduction of RecA, a recombination protein controlled by LexA, does not substitute. Second, we provide evidence that RecF protein is required for efficient mutation in its SOS-inducing capacity. This implies that the DNA signal provoking SOS during adaptive mutation is not a DNA double-strand break (DSB) as postulated previously (e.g., ref. 18), and implies that there are ssDNA intermediates in mutation other than at DSBs. Third, we find evidence of an SOS-controlled repressor of adaptive mutation, PsiB, a protein known to inhibit RecA* activity. The adaptive mutation response appears to occur within a narrow window in the continuum of levels of SOS induction. These results (i) indicate that adaptive mutation is a tightly regulated response, (ii) identify part of the signal transduction pathway that controls it, and (iii) illuminate possible DNA intermediates in that signal transduction pathway.
If you don't think mutational hotspots are hotspots because they are difficult to replicate with 100% fidelity, how do YOU explain what makes them “hot” for mutation?
A computing cell.
Evolutionary Potential of the ebgA Gene
The wild-type enzyme, whose function in nature is unknown, is a very feeble P-galactosidase whose activity toward lactose (galactosyl-P- 1,4-D-glucose) and its analog lactulose (galactosyl-B- 1,4-D-fructose) is so ineffective that, in AZacZ strains, those sugars cannot be utilized for growth even when the operon is expressed constitutively (ebgR_) at a level such that EBG enzyme constitutes 5% of the cell’s soluble protein (Hall 1982). When a AZacZ ebgR - strain is subjected to selection for growth on lactose, only two phenotypic classes of singlestep spontaneous mutants have been obtained among several hundred independent isolates. Class I mutants grow rapidly on lactose, but fail to grow on lactulose, as the result of a mutation that increases the catalytic ef-ficiency of EBG enzyme for lactose 40-fold but does not increase its efficiency for lactulose ( Hall 198 1) . Class II mutants grow rapidly on lactulose, but only moderately well on lactose, as the result of a mutation that increases the catalytic efficiency of EBG enzyme for lactulose 48- fold but increases its efficiency for lactose only IO-fold (Hall 198 1). Selection for growth on lactulose yields only Class II mutants. Class I mutants could not be genetically resolved from each other (i.e., they mapped to the same site), and the biochemical properties of EBG enzyme encoded by Class I mutants were indistinguishable from each other. Similarly, Class II mutants were genetically and biochemically indistinguishable from each other. There was about 1% recombination between the sites of Class I and Class II mutations (Hall and Zuzel 1980b).
The ebgA-encoded a-subunit of EBG enzyme shares 35.5% sequence identity with the la&encoded P-galactosidase (Hall et al. 1989); thus the ebg operon is clearly paralogous with the Zac operon. The crystal structure of lad P-galactosidase has recently been solved (Jacobson et al. 1994)) and the “active-site” amino acids that form the substrate pocket have been identified. The homology between the two proteins is underscored by the observation that 13 of the 15 active-site residues so identified are conserved between the ebg a-peptide and the ZacZ P-galactosidase.
The evolutionary potential of the organism, E. coli K 12, for evolving lactose utilization appears to be limited to the ebg operon. Despite extensive efforts, a strain that carried deletions both in lad and in ebgA was unable to generate spontaneous, chemically induced, or UVinduced mutants that were able to utilize lactose (B. Hall, unpublished observations; D. Hartl, personal communication ) .
170
posted on
06/12/2009 5:09:53 PM PDT
by
AndrewC
(Metanoia)
To: allmendream
“So back to the subject of your supposed constraint. What would you imagine would be the constraint on a mouse's DNA that would prevent it evolving into something like a rat?” [excerpt]
Thats a different subject.
To suggest that its not is disingenuous.
171
posted on
06/12/2009 5:17:06 PM PDT
by
Fichori
To: AndrewC
You are right that sometimes it is...
A molecule hits a receptor that activates a transcriptional repressor that turns off a gene.
Nowhere is ther an intelligence choice made. The entire system was primed to recieve a signal that will elicit a response through MOLECULAR CHAIN REACTIONS.
You think the cell is making choices rather than responding to molecular stimulus? The molecules sure seem to be determining the outcomes wherever we look at what these biological molecules are doing.
And then you cite a paper talking about how mutations can improve the catalytic efficiency of enzymes?
Yep, it sure can, once again illustrating why a bacteria would want to express error prone DNA polymerase in order to increase its rate of evolution.
172
posted on
06/12/2009 5:19:17 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: Fichori
It is the same subject.
How is it different?
Are you suggesting there are constraints on a human’s DNA that are not on a mouse's DNA?
173
posted on
06/12/2009 5:20:06 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: allmendream
“It is the same subject.” [excerpt]
You have defined it as the same.
Which is worth absolutely nothing.
“Are you suggesting there are constraints on a human’s DNA that are not on a mouse's DNA?” [excerpt]
Still trying the old bait-n-switch I see.
174
posted on
06/12/2009 5:25:21 PM PDT
by
Fichori
To: Fichori; allmendream; CottShop
“The availability of the chimp genome sequence in 2005 has provided a more realistic comparison. It should be noted that the chimp genome was sequenced to a much less stringent level than the human genome, and when completed it initially consisted of a large set of small un-oriented and random fragments. To assemble these DNA fragments into contiguous sections that represented large regions of chromosomes, the human genome was used as a guide or framework to anchor and orient the chimp sequence. Thus, the evolutionary assumption of a supposed ape to human transition was used to assemble the otherwise random chimp genome.
Comparing chimp data that has been arranged to look like human data, to human data is a dishonest card trick.”
—Such a thing is not done because of “evolutionary assumptions” or to make chimp dna look like human dna or any such thing - it’s done simply because that’s the common sense way to compare dna. The same thing is done when comparing the dna of various humans.
For example... suppose we have 2 people and we want to compare their dna. The dna of “Bill” is mapped, and then the dna of “Bob” is mapped. The results of all the chromosomes for both people are put into 2 strings roughly 3 billion letters long (atcggtaccat... etc) each. We then compare the 2 strings.
The results show that the 2 are exactly alike... all 3 billion letters... except that Bob has one less nucleotide - at the start of the first chromosome. That means if you line the two strings up, the result will be that they are 0% alike - because everything is shifted by one because of the missing nucleotide. So 1 letter is different out of 3 billion and they are 0% alike! Obviously, if you want to have a meaningful comparison, you shift the dna string of Bob up one.
That wasn’t just a hypothetical, in each of us are insertions and deletions and copying. It’s not even uncommon for large chunks of (tens of million of nucleotides) to be in a different place among 2 different people - sometimes a piece of one chromosome will break off and connect to another. This all has to be accounted for when comparing dna - it’s not easy.
So it should be noted that when the comparison of chimp and human dna is done, it’s a comparison of of *A* human and *A* chimp, and they were compared precisely the same we compare ANY dna.
As for how similar chimp and human dna are, I’d use the figure of 96%, as this site suggests:
http://www.genome.gov/15515096
“The DNA sequence that can be directly compared between the two genomes is almost 99 percent identical. When DNA insertions and deletions are taken into account, humans and chimps still share 96 percent of their sequence.”
It’s also untrue that geneticists are concentrating on the areas where the 2 genomes are most similar.
How many geneticists do you think are out that want to find a pivotal difference between humans and other animals? Finding differences are the “Eureka!” moments.
To: goodusername
Comparing chimp data that has been arranged to look like human data, to human data is a dishonest card trick.
“—Such a thing is not done because of “evolutionary assumptions” or to make chimp dna look like human dna or any such thing - it’s done simply because that’s the common sense way to compare dna. The same thing is done when comparing the dna of various humans.” [excerpt]
Objectivity.
You ain't got it.
176
posted on
06/12/2009 5:49:10 PM PDT
by
Fichori
To: AndrewC
In your mind. Reread the post. I gave the universe of possibilities, then I specified a particular sequence. I then specified a method to generate that particular sequence. What you describe is the probability of shuffling a sequence which is one, certainty. Like this... No. I showed you that it is possible to get the sequence with the same odds as you describe. Ouch! It happed again. Imagine that!
To: AndrewC
The chance of tossing neither is zero:WRONG!It is possible for it to land on its edge!
To: Fichori
So what is the constraint on EITHER a mouse or a human’s DNA that would keep them from changing their genetic DNA by a few % into another perfectly functional gene?
Your entire premise is that somehow mutation is constrained and that would prevent different species being descended from a common ancestor. What is the nature of the constraint?
179
posted on
06/12/2009 5:55:34 PM PDT
by
allmendream
("Wealth is EARNED not distributed, so how could it be redistributed?")
To: allmendream
And then you cite a paper talking about how mutations can improve the catalytic efficiency of enzymes?But of course you leave out the fact that there are only two mutations in a specific enzyme whose function is not known. That is not accidental. They tried to generate the activity by deleting both genes capable of producing the desired activity with no success. You also fail to point out that the two genes being discussed are very similar. The mutations are targeted to two specific locations on one specific gene.
A molecule hits a receptor that activates a transcriptional repressor that turns off a gene.
No, again you simplify.
To summarize: (i) PsiB appears to inhibit adaptive mutation when the LexA regulon is constitutively de-repressed in a lexADef mutant; and (ii) two proteins that modulate RecA* activity, DinI and PsiB, affect adaptive mutation positively and negatively. These data suggest that RecA* activity is critical in adaptive mutation, that if RecA* activity is either too high or too low, mutation is decreased. These results indicate a tight control over adaptive mutation by factors modulating the SOS response, and provide evidence of SOS regulation of adaptive mutation independent of particular LexA alleles.
180
posted on
06/12/2009 6:01:33 PM PDT
by
AndrewC
(Metanoia)
Navigation: use the links below to view more comments.
first previous 1-20 ... 141-160, 161-180, 181-200 ... 261-271 next last
Disclaimer:
Opinions posted on Free Republic are those of the individual
posters and do not necessarily represent the opinion of Free Republic or its
management. All materials posted herein are protected by copyright law and the
exemption for fair use of copyrighted works.
FreeRepublic.com is powered by software copyright 2000-2008 John Robinson
 . The σ-algebra
. The σ-algebra  contains
contains  events, namely,
events, namely,  : heads,
: heads,  : tails,
: tails, 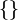 : neither heads nor tails, and
: neither heads nor tails, and  : heads or tails. So,
: heads or tails. So,  There is a fifty percent chance of tossing either heads or tail:
There is a fifty percent chance of tossing either heads or tail:  thus
thus  The chance of tossing neither is zero:
The chance of tossing neither is zero:  and the chance of tossing one or the other is one:
and the chance of tossing one or the other is one: 