Posted on 02/08/2005 3:50:43 AM PST by PatrickHenry
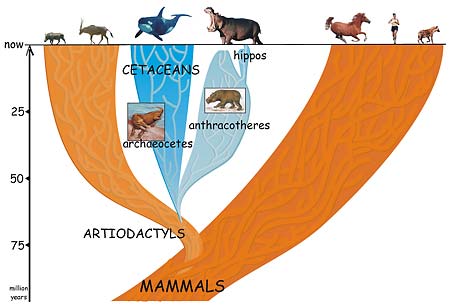
A group of four-footed mammals that flourished worldwide for 40 million years and then died out in the ice ages is the missing link between the whale and its not-so-obvious nearest relative, the hippopotamus.
The conclusion by University of California, Berkeley, post-doctoral fellow Jean-Renaud Boisserie and his French colleagues finally puts to rest the long-standing notion that the hippo is actually related to the pig or to its close relative, the South American peccary. In doing so, the finding reconciles the fossil record with the 20-year-old claim that molecular evidence points to the whale as the closest relative of the hippo.
"The problem with hippos is, if you look at the general shape of the animal it could be related to horses, as the ancient Greeks thought, or pigs, as modern scientists thought, while molecular phylogeny shows a close relationship with whales," said Boisserie. "But cetaceans – whales, porpoises and dolphins – don't look anything like hippos. There is a 40-million-year gap between fossils of early cetaceans and early hippos."
In a paper appearing this week in the Online Early Edition of the Proceedings of the National Academy of Sciences, Boisserie and colleagues Michel Brunet and Fabrice Lihoreau fill in this gap by proposing that whales and hippos had a common water-loving ancestor 50 to 60 million years ago that evolved and split into two groups: the early cetaceans, which eventually spurned land altogether and became totally aquatic; and a large and diverse group of four-legged beasts called anthracotheres. The pig-like anthracotheres, which blossomed over a 40-million-year period into at least 37 distinct genera on all continents except Oceania and South America, died out less than 2 and a half million years ago, leaving only one descendent: the hippopotamus.
This proposal places whales squarely within the large group of cloven-hoofed mammals (even-toed ungulates) known collectively as the Artiodactyla – the group that includes cows, pigs, sheep, antelopes, camels, giraffes and most of the large land animals. Rather than separating whales from the rest of the mammals, the new study supports a 1997 proposal to place the legless whales and dolphins together with the cloven-hoofed mammals in a group named Cetartiodactyla.
"Our study shows that these groups are not as unrelated as thought by morphologists," Boisserie said, referring to scientists who classify organisms based on their physical characteristics or morphology. "Cetaceans are artiodactyls, but very derived artiodactyls."

The origin of hippos has been debated vociferously for nearly 200 years, ever since the animals were rediscovered by pioneering French paleontologist Georges Cuvier and others. Their conclusion that hippos are closely related to pigs and peccaries was based primarily on their interpretation of the ridges on the molars of these species, Boisserie said.
"In this particular case, you can't really rely on the dentition, however," Boisserie said. "Teeth are the best preserved and most numerous fossils, and analysis of teeth is very important in paleontology, but they are subject to lots of environmental processes and can quickly adapt to the outside world. So, most characteristics are not dependable indications of relationships between major groups of mammals. Teeth are not as reliable as people thought."
As scientists found more fossils of early hippos and anthracotheres, a competing hypothesis roiled the waters: that hippos are descendents of the anthracotheres.
All this was thrown into disarray in 1985 when UC Berkeley's Vincent Sarich, a pioneer of the field of molecular evolution and now a professor emeritus of anthropology, analyzed blood proteins and saw a close relationship between hippos and whales. A subsequent analysis of mitochondrial, nuclear and ribosomal DNA only solidified this relationship.
Though most biologists now agree that whales and hippos are first cousins, they continue to clash over how whales and hippos are related, and where they belong within the even-toed ungulates, the artiodactyls. A major roadblock to linking whales with hippos was the lack of any fossils that appeared intermediate between the two. In fact, it was a bit embarrassing for paleontologists because the claimed link between the two would mean that one of the major radiations of mammals – the one that led to cetaceans, which represent the most successful re-adaptation to life in water – had an origin deeply nested within the artiodactyls, and that morphologists had failed to recognize it.
This new analysis finally brings the fossil evidence into accord with the molecular data, showing that whales and hippos indeed are one another's closest relatives.
"This work provides another important step for the reconciliation between molecular- and morphology-based phylogenies, and indicates new tracks for research on emergence of cetaceans," Boisserie said.
Boisserie became a hippo specialist while digging with Brunet for early human ancestors in the African republic of Chad. Most hominid fossils earlier than about 2 million years ago are found in association with hippo fossils, implying that they lived in the same biotopes and that hippos later became a source of food for our distant ancestors. Hippos first developed in Africa 16 million years ago and exploded in number around 8 million years ago, Boisserie said.
Now a post-doctoral fellow in the Human Evolution Research Center run by integrative biology professor Tim White at UC Berkeley, Boisserie decided to attempt a resolution of the conflict between the molecular data and the fossil record. New whale fossils discovered in Pakistan in 2001, some of which have limb characteristics similar to artiodactyls, drew a more certain link between whales and artiodactyls. Boisserie and his colleagues conducted a phylogenetic analysis of new and previous hippo, whale and anthracothere fossils and were able to argue persuasively that anthracotheres are the missing link between hippos and cetaceans.
While the common ancestor of cetaceans and anthracotheres probably wasn't fully aquatic, it likely lived around water, he said. And while many anthracotheres appear to have been adapted to life in water, all of the youngest fossils of anthracotheres, hippos and cetaceans are aquatic or semi-aquatic.
"Our study is the most complete to date, including lots of different taxa and a lot of new characteristics," Boisserie said. "Our results are very robust and a good alternative to our findings is still to be formulated."
Brunet is associated with the Laboratoire de Géobiologie, Biochronologie et Paléontologie Humaine at the Université de Poitiers and with the Collège de France in Paris. Lihoreau is a post-doctoral fellow in the Département de Paléontologie of the Université de N'Djaména in Chad.
The work was supported in part by the Mission Paléoanthropologique Franco-Tchadienne, which is co-directed by Brunet and Patrick Vignaud of the Université de Poitiers, and in part by funds to Boisserie from the Fondation Fyssen, the French Ministère des Affaires Etrangères and the National Science Foundation's Revealing Hominid Origins Initiative, which is co-directed by Tim White and Clark Howell of UC Berkeley.
I bet the first whale that jumped on the beach and suddenly started breathing air only did so to get away from all of his pals and their cruel jokes about his freakish half hippo appearance.
pong
"I assure you, all of our ancestors were fully human."
I assure you, you are wrong. Evolution is a fact.
Another candidate for remedial biology.
ALL whales breathe air.
There you have it - if given enough time my 65 Ford Mustang could evolve to be a fruit fly. They say "life" somehow sprang from un-life so it'll eventually work out the problems.
Ummmm....
You've got things a bit reversed. Land animals didn't evolve from whales. Whales evolved from land animals.
Or did they?? hmmmm
yes they did is the answer
Scientists find missing link between Janeane Garofolo and her nearest relative Roseanne Barr.
Both are fat and unfunny.
nice to see genetics beginning to take over where morphological taxonomy drops the ball.
Who are "they?" How's that deception thing working out for you?

whale

Hippo
for later
Yep. Whales evolved from primitive land whales (aka pakiceti) that looked something like this:

No they werent. Here are just a few of the many things you're extremely ignorant of on this topic:
(From Plagiarized Errors and Molecular Genetics)That's just a quick layman-level overview of *one* of the many ways that whale evolution has been verified. For more technical examinations along several independent lines of evidence, see for example:.
A particularly impressive example of shared retroposons has recently been reported linking cetaceans (whales, dolphins and porpoises) to ruminants and hippopotamuses, and it is instructive to consider this example in some detail. Cetaceans are sea-living animals that bear important similarities to land-living mammals; in particular, the females have mammary glands and nurse their young. Scientists studying mammalian anatomy and physiology have demonstrated greatest similarities between cetaceans and the mammalian group known as artiodactyls (even-toed ungulates) including cows, sheep, camels and pigs. These observations have led to the evolutionist view that whales evolved from a four-legged artiodactyl ancestor that lived on land. Creationists have capitalized on the obvious differences between the familiar artiodactyls and whales, and have ridiculed the idea that whales could have had four-legged land-living ancestors. Creationists who claim that cetaceans did not arise from four-legged land mammals must ignore or somehow dismiss the fossil evidence of apparent whale ancestors looking exactly like one would predict for transitional species between land mammals and whales--with diminutive legs and with ear structures intermediate between those of modern artiodactyls and cetaceans (Nature 368:844,1994; Science 263: 210, 1994). (A discussion of fossil ancestral whale species with references may be found at http://www.talkorigins.org/faqs/faq-transitional/part2b.html#ceta) Creationists must also ignore or dismiss the evidence showing the great similarity between cetacean and artiodactyl gene sequences (Molecular Biology & Evolution 11:357, 1994; ibid 13: 954, 1996; Gatesy et al, Systematic Biology 48:6, 1999).
Recently retroposon evidence has solidified the evolutionary relationship between whales and artiodactyls. Shimamura et al. (Nature 388:666, 1997; Mol Biol Evol 16: 1046, 1999; see also Lum et al., Mol Biol Evol 17:1417, 2000; Nikaido and Okada, Mamm Genome 11:1123, 2000) studied SINE sequences that are highly reduplicated in the DNA of all cetacean species examined. These SINES were also found to be present in the DNA of ruminants (including cows and sheep) but not in DNA of camels and pigs or more distantly related mammals such as horse, elephant, cat, human or kangaroo. These SINES apparently originated in a specific branch of ancestral artiodactyls after this branch diverged from camels, pigs and other mammals, but before the divergence of the lines leading to modern cetaceans, hippopotamus and ruminants. (See Figure 5.) In support of this scenario, Shimamura et al. identified two specific insertions of these SINES in whale DNA (insertions B and C in Figure 5) and showed that in DNA of hippopotamus, cow and sheep these same two sites contained the SINES; but in camel and pig DNA the same sites were "empty" of insertions. More recently, hippopotamus has been identified as the closest living terrestrial relative of cetaceans since hippos and whales share retroposon insertions (illustrated by D and E in Figure 5) that are not found in any other artiodactyls (Nikaido et al, PNAS 96:10261, 1999). The close hippo-whale relationship is consistent with previously reported sequence similarity comparisons (Gatesy, Mol Biol Evol 14:537, 1997) and with recent fossil finds (Gingerich et al., Science 293:2239, 2001; Thewissen et al., Nature 413:277, 2001) that resolve earlier paleontological conflicts with the close whale-hippo relationship. (Some readers have wondered: if ruminants are more closely related to whales than to pigs and camels, why are ruminants anatomically more similar to pigs and camels than they are to whales? Apparently this results from the fact that ruminants, pigs and camels changed relatively little since their last common ancestor, while the cetacean lineage changed dramatically in adapting to an aquatic lifestyle, thereby obliterating many of the features -- like hooves, fur and hind legs -- that are shared between its close ruminant relatives and the more distantly related pigs and camels. This scenario illustrates the fact that the rapid evolutionary development of adaptations to a new niche can occur through key functional mutations, leaving the bulk of the DNA relatively unchanged. The particularly close relationship between whales and hippos is consistent with several shared adaptations to aquatic life, including use of underwater vocalizations for communication and the absence of hair and sebaceous glands.) Thus, retroposon evidence strongly supports the derivation of whales from a common ancestor of hippopotamus and ruminants, consistent with the evolutionary interpretation of fossils and overall DNA sequence similarities. Indeed, the logic of the evidence from shared SINEs is so powerful that SINEs may be the best available characters for deducing species relatedness (Shedlock and Okada, Bioessays 22:148, 2000), even if they are not perfect (Myamoto, Curr. Biology 9:R816, 1999).
Figure 5. Specific SINE insertions can act as "tracers" that illuminate phylogenetic relationships. This figure summarizes some of the data on SINEs found in living artiodactyls and shows how the shared insertions can be interpreted in relation to evolutionary branching. A specific SINE insertion event ("A" in the Figure) apparently occurred in a primitive common ancestor of pigs, ruminants, hippopotamus and cetaceans, since this insertion is present in these modern descendants of that common ancestor; but it is absent in camels, which split off from the other species before this SINE inserted. More recent insertions B and C are present only in ruminants, hippopotamus and cetaceans. Insertions D and E are shared only by hippopotamus and cetaceans, thereby identifying hippopotamus as the closest living relative of cetaceans (at least among the species examined in these studies). SINE insertions F and G occurred in the ruminant lineage after it diverged from the other species; and insertions H and I occurred after divergence of the cetacean lineage.
SINE Evolution, Missing Data, and the Origin of WhalesAnd much, much more.Evidence from Milk Casein Genes that Cetaceans are Close Relatives of Hippopotamid Artiodactyls
Analyses of mitochondrial genomes strongly support a hippopotamus±whale clade
A new Eocene archaeocete (Mammalia, Cetacea) from India and the time of origin of whales
Mysticete (Baleen Whale) Relationships Based upon the Sequence of the Common Cetacean DNA Satellite1
Eocene evolution of whale hearing
Novel Phylogeny of Whales Revisited but Not Revised
New Morphological Evidence for the Phylogeny of Artiodactyla, Cetacea, and Mesonychidae
So, when you say, "Scientists now theorize that it evolved from a hippo-like creature, though they really have no idea", I only have one question for you: Are you outright lying, or are you just monumentally ignorant (but still arrogant enough to spout off about something you actually know so little about)?
That's not a rhetorical question. Please respond.
So unless you can elucidate some opposing scenario in which all of the above findings are *better* accounted for than in the evolutionary scenario, go run off and play with the other creationists -- the evolutionary biologists are doing adult-type stuff that you apparently can't grasp, and have no real interest in learning about.
Are there any other theories that has promoted as much searching for evidence to support it than evolution?
No need to "search" for the evidence for evolution, it arrives in a flood every time anything even remotely tangential to life is examined. For example:
There's plenty more where that came from -- let me know when you're done reading those and I'll give you another ten thousand or so.The Evolution of Improved Fitness by random mutation plus selection
Ancient Jumping DNA May Have Evolved Into Key Component Of Human Immune System
Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system
New insights into V(D)J recombination and its role in the evolution of the immune system
Evolution and developmental regulation of the major histocompatibility complex
Evolution of the IL-6/class IB cytokine receptor family in the immune and nervous systems
Development of an immune system
The ancestor of the adaptive immune system was the CAM system for organogenesis
The evolutionary origins of immunoglobulins and T-cell receptors: possibilities and probabilities
Evolutionary perspectives on amyloid and inflammatory features of Alzheimer disease
Organization of the human RH50A gene (RHAG) and evolution of base composition of the RH gene family.
Molecular evolution of the vertebrate immune system.
Morphostasis: an evolving perspective.
Rapid evolution of immunoglobulin superfamily C2 domains expressed in immune system cells.
Evolutionary assembly of blood coagulation proteins
Exon and domain evolution in the proenzymes of blood coagulation and fibrinolysis
Evolution of proteolytic enzymes
Evolution of vertebrate fibrin formation and the process of its dissolution.
Common Parasite Overturns Traditional Beliefs About The Evolution And Role Of Hemoglobin
Scientists Discover How Bacteria Protect Themselves Against Immune System
Reduction of two functional gamma-globin genes to one: an evolutionary trend in New World monkeys
Evolutionary history of introns in a multidomain globin gene
Hemoglobin A2: origin, evolution, and aftermath
Early evolution of microtubules and undulipodia
Flagellar beat patterns and their possible evolution
The evolutionary origin and phylogeny of eukaryote flagella
Dynein family of motor proteins: present status and future questions
Origins of the nucleate organisms
The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella
The evolution of cellular movement in eukaryotes: the role of microfilaments and microtubules
Kinesin Motor Phylogenetic Tree
Evolution of a dynamic cytoskeleton
Isolation, characterization and evolution of nine pufferfish (Fugu rubripes) actin genes
Evolution of chordate actin genes: evidence from genomic organization and amino acid sequences
Co-evolution of ligand-receptor pairs in the vasopressin/oxytocin superfamily of bioactive peptides
The evolution of the synapses in the vertebrate central nervous system
Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria.
The evolution of metabolic cycles
Evolution of the first metabolic cycles
Speculations on the origin and evolution of metabolism
The Molecular Anatomy of an Ancient Adaptive Event
Biochemical pathways in prokaryotes can be traced backward through evolutionary time
Enzyme specialization during the evolution of amino acid biosynthetic pathways
Enzyme recruitment in evolution of new function
Bioenergetics: the evolution of molecular mechanisms and the development of bioenergetic concepts
Theoretical approaches to the evolutionary optimization of glycolysis--chemical analysis
The evolution of kinetoplastid glycosomes
Stepwise molecular evolution of bacterial photosynthetic energy conversion
Evolution of photosynthetic reaction centers and light harvesting chlorophyll proteins
Evolution of photosynthetic reaction centers
Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins
Evolution of the control of pigment and plastid development in photosynthetic organisms
Chemical evolution of photosynthesis
Molecular evolution of ruminant lysozymes
Evolution of stomach lysozyme: the pig lysozyme gene
Molecular basis for tetrachromatic color vision
Molecular evolution of the Rh3 gene in Drosophila
The evolution of rhodopsins and neurotransmitter receptors
Optimization, constraint, and history in the evolution of eyes
A pessimistic estimate of the time required for an eye to evolve
The eye of the blind mole rat (Spalax ehrenbergi): regressive evolution at the molecular level
Programming the Drosophila embryo
Evolution of chordate hox gene clusters
Hox genes in brachiopods and priapulids and protostome evolution.
Radical evolutionary change possible in a few generations
Evolution Re-Sculpted Animal Limbs By Genetic Switches Once Thought Too Drastic For Survival
The origin and evolution of animal appendages
Hox genes in evolution: protein surfaces and paralog groups
Evolution of the insect body plan as revealed by the Sex combs reduced expression pattern
Sea urchin Hox genes: insights into the ancestral Hox cluster
Theoretical approaches to the analysis of homeobox gene evolution
Teleost HoxD and HoxA genes: comparison with tetrapods and functional evolution of the HOXD complex
Evolutionary origin of insect wings from ancestral gills
Tracing backbone evolution through a tunicate's lost tail
The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster.
Gene duplications in evolution of archaeal family B DNA polymerases
Adaptive amino acid replacements accompanied by domain fusion in reverse transcriptase
Molecular evolution of genes encoding ribonucleases in ruminant species
Studies on the sites expressing evolutionary changes in the structure of eukaryotic 5S ribosomal RNA
Evolution of a Transfer RNA Gene Through a Point Mutation in the Anticodon
Universally conserved translation initiation factors
Genetic code in evolution: switching species-specific aminoacylation with a peptide transplant
Evolution of transcriptional regulatory elements within the promoter of a mammalian gene.
Codon reassignment and amino acid composition in hemichordate mitochondria.
Determining divergence times of the major kingdoms of living organisms with a protein clock
The multiplicity of domains in proteins
Characterization, primary structure, and evolution of lamprey plasma albumin
The origins and evolution of eukaryotic proteins
Evolution of vertebrate fibrin formation and the process of its dissolution.
Vastly Different Virus Families May Be Related
Selective sweep of a newly evolved sperm-specific gene in Drosophila
Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions
Molecular evolution of the histidine biosynthetic pathway
Accelerated evolution in inhibitor domains of porcine elafin family members
Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod
Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish
Evolution of an antifreeze glycoprotein
A model for the evolution of the plastid sec apparatus inferred from secY gene phylogeny
The evolutionary history of the amylase multigene family in Drosophila pseudoobscura
The evolution of an allosteric site in phosphorylase
Molecular evolution of fish neurohypophysial hormones: neutral and selective evolutionary mechanisms
Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function?
Evolutionary relationships of the carbamoylphosphate synthetase genes
The molecular evolution of the small heat-shock proteins in plants
Evolutionary history of the 11p15 human mucin gene family.
Molecular evolution of the aldo-keto reductase gene superfamily.
A Classification of Possible Routes of Darwinian Evolution
Generation of evolutionary novelty by functional shift
Mobile DNA Sequences Could Be The Cause Of Chromosomal Mutations During The Evolution Of Species
Minor Shuffle Makes Protein Fold
Genetic Stowaways May Contribute To Evolutionary Change
Evolutionary Molecular Mechanism In Mammals Found
Cases of ancient mobile element DNA insertions that now affect gene regulation
Punctuated evolution caused by selection of rare beneficial mutations
The origin of programmed cell death
The origin and early development of biological catalysts
DNA secondary structures and the evolution of hypervariable tandem arrays
Episodic adaptive evolution of primate lysozymes
Genome plasticity as a paradigm of eubacteria evolution
Evolutionary motif and its biological and structural significance
Exon shuffling and other ways of module exchange
New Drosophila introns originate by duplication.
Evolution and the structural domains of proteins
The role of constrained self-organization in genome structural evolution
The coevolution of gene family trees
The evolution of metabolic cycles
The emergence of major cellular processes in evolution
A hardware interpretation of the evolution of the genetic code
Speculations on the origin and evolution of metabolism
Probabilistic reconstruction of ancestral protein sequences
The contribution of slippage-like processes to genome evolution
Molecular evolution in bacteria
The structural basis of molecular adaptation.
Mitochondrial DNA: molecular fossils in the nucleus
Cases of ancient mobile element DNA insertions that now affect gene regulation
Tiggers and DNA transposon fossils in the human genome
The eye of the blind mole rat (Spalax ehrenbergi): regressive evolution at the molecular level
Tiggers and DNA transposon fossils in the human genome
Gene competition and the possible evolutionary role of tumours
New Scientist Planet Science: Replaying life
Molecular evolution of an arsenate detoxification pathway by DNA shuffling
UB Researcher Developing Method That Employs Evolution To Develop New Drug Leads
Exploring the functional robustness of an enzyme by in vitro evolution
Evolutionary algorithms in computer-aided molecular design
Evolution of Enzymes for the Metabolism of New Chemical Inputs into the Environment
Evolution of Amino Acid Metabolism Inferred through Cladistic Analysis
Integrating the Universal Metabolism into a Phylogenetic Analysis
Serial segmental duplications during primate evolution result in complex human genome architecture
Phylogeny determined by protein domain content
Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily
Molecular archaeology of the Escherichia coli genome
Comparative Genomics of the Eukaryotes
Asymmetric Sequence Divergence of Duplicate Genes
The Genetic Core of the Universal Ancestor
Evolutionary History of Chromosome 20
Reconstructing large regions of an ancestral mammalian genome in silico
Occurrence and Consequences of Coding Sequence Insertions and Deletions in Mammalian Genomes
The Origin of Human Chromosome 1 and Its Homologs in Placental Mammals
On the RNA World: Evidence in Favor of an Early Ribonucleopeptide World
Inhibition of Ribozymes by Deoxyribonucleotides and the Origin of DNA
They're really stretching here.
No, you're really in denial.
When a species had to develop a defense measure to keep from being wiped out by predators this susposedly took millions of years, they must have been in hiding wating for armor, wings, poison glands or whatever to develop.
This cartoonish scenario of yours reveals a remarkable ignorance of evolutionary biology. Wherever you went to school, you should sue.
I assure you, all of our ancestors were fully human.
I assure you, you have no idea what you're talking about. Your statement clearly indicates that you haven't eve bothered to look -- there is a *massive* amount of evidence supporting common descent via evolution. In fact, there are several dozen *independent* lines of evidence that support evolution -- how do you explain your ignorance on this topic, and your willingness to make absolute claims about it without knowing the first thing about it?
For example, here's one from a previous post of mine:
Or how about:Background: Retroviruses reproduce by entering a cell of a host (like, say, a human), then embedding their own viral DNA into the cell's own DNA, which has the effect of adding a "recipe" for manufacturing more viruses to the cell's "instruction book". The cell then follows those instructions because it has no reason (or way) to "mistrust" the DNA instructions it contains. So the virus has converted the cell into a virus factory, and the new viruses leave the cell, and go find more cells to infect, etc.
However, every once in a while a virus's invasion plans don't function exactly as they should, and the virus's DNA (or portions of it) gets embedded into the cell's DNA in a "broken" manner. It's stuck into there, becoming part of the cell's DNA, but it's unable to produce new viruses. So there it remains, causing no harm. If this happens in a regular body cell, it just remains there for life as a "fossil" of the past infection and goes to the grave with the individual it's stuck in. All of us almost certainly contain countless such relics of the past viral infections we've fought off.
However... By chance this sometimes happens to a special cell in the body, a gametocyte cell that's one of the ones responsible for making sperm in males and egg cells in females, and if so subsequent sperm/eggs produced by that cell will contain copies of the "fossil" virus, since now it's just a portion of the entire DNA package of the cell. And once in a blue moon such a sperm or egg is lucky enough to be one of the few which participate in fertilization and are used to produce a child -- who will now inherit copies of the "fossilized" viral DNA in every cell of his/her body, since all are copied from the DNA of the original modified sperm/egg.
So now the "fossilized" viral DNA sequence will be passed on to *their* children, and their children's children, and so on. Through a process called neutral genetic drift, given enough time (it happens faster in smaller populations than large) the "fossil" viral DNA will either be flushed out of the population eventually, *or* by luck of the draw end up in every member of the population X generations down the road. It all depends on a roll of the genetic dice.
Due to the hurdles, "fossil" retroviral DNA strings (known by the technical name of "endogenous retroviruses") don't end up ubiquitous in a species very often, but it provably *does* happen. In fact, the Human DNA project has identified literally *thousands* of such fossilized "relics" of long-ago ancestral infections in the human DNA.
And several features of these DNA relics can be used to demonstrate common descent, including their *location*. The reason is that retroviruses aren't picky about where their DNA gets inserted into the host DNA. Even in an infection in a *single* individual, each infected cell has the retroviral DNA inserted into different locations than any other cell. Because the host DNA is so enormous (billions of basepairs in humans, for example), the odds of any retroviral insertion event matching the insertion location of any other insertion event are astronomically low. The only plausible mechanism by which two individuals could have retroviral DNA inserted into exactly the same location in their respective DNAs is if they inherited copies of that DNA from the same source -- a common ancestor.
Thus, shared endogenous retroviruses between, say, ape and man is almost irrefutable evidence that they descended from a common ancestor. *Unless* you want to suggest that they were created separately, and then a virus they were both susceptible to infected both a man and an ape in EXACTLY the same location in their DNAs (the odds of such a match by luck are literally on the order of 1,000,000,000,000,000,000 to 1...), *and* that the infections both happened in their gametocyte cells (combined odds on the order of 1,000,000 to 1) *and* that the one particular affected gametocyte is the one which produces the egg or sperm which is destined to produce an offspring (*HUGE* odds against), and *then* the resulting modified genome of the offspring becomes "fixed" in each respective population (1 out of population_size^squared)...
Then repeat that for *each* shared endogenous retrovirus (there are many) you'd like to claim was acquired independently and *not* from a shared ancestor...
Finally, you'd have to explain why, for say species A, B, and C, the pattern of shared same-location retroviruses is always *nested*, never *overlapped*. For example, all three will share some retroviruses, then A and B will both share several more, but if so then B *never* shares one with C that A doesn't also have (or at least remnants of).
In your "shared infection due to genetic similarities" suggestion, even leaving aside the near statistical impossibility of the infections leaving genetic "scars" in *exactly* the same locations in independent infections, one would expect to find cases of three species X, Y, and Z, where the degree of similarity was such that Y was "between" X and Z on some similarity scale, causing the same disease to befall X and Y but not Z, and another disease to affect Y and Z but not X. And yet, we don't find this in genetic markers. The markers are found in nested sequence, which is precisely what we would expect to see in cases of inheritance from common ancestry.
Here, for example, is an ancestry tree showing the pattern of shared same-location endogenous retroviruses of type HERV-K among primates:
This is just a partial list for illustration purposes -- there are many more.
Each labeled arrow on the chart shows an ERV shared in common by all the branches to the right, and *not* the branches that are "left-and-down". This is the pattern that common descent would make. And common descent is the *only* plausible explanation for it. Furthermore, similar findings tie together larger mammal groups into successively larger "superfamilies" of creatures all descended from a common ancestor.
Any presumption of independent acquisition is literally astronomically unlikely. And "God chose to put broken relics of viral infections that never actually happened into our DNA and line them up only in patterns that would provide incredibly strong evidence of common descent which hadn't actually happened" just strains credulity (not to mention would raise troubling questions about God's motives for such a misleading act).
Once again, the evidence for common descent -- as opposed to any other conceivable alternative explanation -- is clear and overwhelming.
Wait, want more? Endogenous retroviruses are just *one* type of genetic "tag" that makes perfect sense evolutionary and *no* sense under any other scenario. In addition to ERV's, there are also similar arguments for the patterns across species of Protein functional redundancies, DNA coding redundancies, shared Processed pseudogenes, shared Transposons (including *several* independent varieties, such as SINEs and LINEs), shared redundant pseudogenes, etc. etc. Here, for example, is a small map of shared SINE events among various mammal groups:
Like ERV's, any scenario which suggests that these shared DNA features were acquired separately strains the laws of probability beyond the breaking point, but they make perfect sense from an evolutionary common-descent scenario. In the above data, it is clear that the only logical conclusion is that, for example, the cetaceans, hippos, and ruminants shared a common ancestor, in which SINE events B and C entered its DNA and then was passed on to its descendants, yet this occurred after the point in time where an earlier common ancestor had given rise both to that species, and to the lineage which later became pigs.
And this pattern (giving the *same* results) is repeated over and over and over again when various kinds of molecular evidence from DNA is examined in detail.
The molecular evidence for evolution and common descent is overwhelming. The only alternative is for creationists to deny the obvious and say, "well maybe God decided to set up all DNA in *only* ways that were consistent with an evolutionary result even though He'd have a lot more options open to him, even including parts which by every measure are useless and exactly mimic copy errors, ancient infections, stutters, and other garbage inherited from nonexistent shared ancestors"...
Humans have 23 pairs of chromosomes ---chimps and gorillas have 24 pairs. How many pairs of chromosomes did the "common ancestor" have? Was it 23 or 24 pairs? How do you "evolve" missing or added chromosomes ---that would happen all at one time.I'm sorry, what was that you were saying about "all of our ancestors were fully human", implying that there is "no evidence" for common ancestry of humans and apes?The common ancestor had 24 chromosomes.
If you look at the gene sequences, you'll find that Chromosome 2 in humans is pretty much just 2 shorter chimpanzee chromosomes pasted end-to-end, with perhaps a slight bit of lost overlap:
(H=Human, C=Chimpanzee, G=Gorilla, O=Orangutan)
Somewhere along the line, after humans split off from the other great apes, or during the split itself, there was an accidental fusion of two chromosomes, end-to-end. Where there used to be 24 chromosomes, now there were 23, but containing the same total genes, so other than a "repackaging", the DNA "instructions" remained the same.
If a chimpanzee gives birth to a creature with 23 chromosomes, that offspring isn't going to be a well-formed chimpanzee able to survive well.
It is if the same genes are present, which they would be in the case of a chromosome fusion.
Evolve would imply the genetic material changes little by little --not some big loss of two chromosomes at once but I don't see how they'd go away gene by gene.
Tacking two chromosomes together end-to-end is not a "big loss" of genes, and it really is a "little by little" change in the total genetic code. It's just been "regrouped" a bit. Instead of coming in 24 "packages", it's now contained in 23, but the contents are the same.
So how, you might ask, would the chromosomes from the first 23-chromosome "fused" individual match up with the 24 chromosomes from its mate when it tried to produce offspring? Very well, thanks for asking. The "top half" of the new extra-long Chromosome 2 would adhere to the original chromosome (call it "2p") from which it was formed, and likewise for the "bottom half" which would adhere to the other original shorter chromosome (call it "2q"). In the picture above, imagine the two chimp chromosomes sliding over to "match up" against the human chromosome. The chimp chromosomes would end up butting ends with each other, or slightly overlapping in a "kink", but chromosomes have overcome worse mismatches (just consider the XY pair in every human male -- the X and the Y chromosome are *very* different in shape, length, and structure, but they still pair up).
In fact, the "rubbing ends" of the matched-up chimp chromosomes, adhering to the double-long human-type chromosome, would be more likely to become fused together themselves.
For studies in which recent chromosome fusions have been discovered and found not to cause infertility, see:
Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Hauffe HC, Searle JB Department of Zoology, University of Oxford, Oxford OX1 3PS, United Kingdom. hauffe@novanet.itIn that last reference, the Przewalski horse, which has 33 chromosomes, and the domestic horse, with 32 chromosomes (due to a fusion), are able to mate and produce fertile offspring.An observed chromosome fusion: Hereditas 1998;129(2):177-80 A new centric fusion translocation in cattle: rob (13;19). Molteni L, De Giovanni-Macchi A, Succi G, Cremonesi F, Stacchezzini S, Di Meo GP, Iannuzzi L Institute of Animal Husbandry, Faculty of Agricultural Science, Milan, Italy.
J Reprod Fertil 1979 Nov;57(2):363-75 Cytogenetics and reproduction of sheep with multiple centric fusions (Robertsonian translocations). Bruere AN, Ellis PM
J Reprod Fertil Suppl 1975 Oct;(23):356-70 Cytogenetic studies of three equine hybrids. Chandley AC, Short RV, Allen WR.
Meanwhile, the question may be asked, how do we know that the human Chromosome 2 is actually the result of a chromsome fusion at/since a common ancestor, and not simply a matter of "different design"?
Well, if two chromsomes accidentally merged, there should be molecular remnants of the original chromosomal structures (while a chromosome designed from scratch would have no need for such leftover "train-wreck" pieces).
Ends of chromosomes have characteristic DNA base-pair sequences called "telomeres". And there are indeed remnants of telomeres at the point of presumed fusion on human Chromosome 2 (i.e., where the two ancestral ape chromosomes merged end-to-end). If I may crib from a web page:
Telomeres in humans have been shown to consist of head to tail repeats of the bases 5'TTAGGG running toward the end of the chromosome. Furthermore, there is a characteristic pattern of the base pairs in what is called the pre-telomeric region, the region just before the telomere. When the vicinity of chromosome 2 where the fusion is expected to occur (based on comparison to chimp chromosomes 2p and 2q) is examined, we see first sequences that are characteristic of the pre-telomeric region, then a section of telomeric sequences, and then another section of pre-telomeric sequences. Furthermore, in the telomeric section, it is observed that there is a point where instead of being arranged head to tail, the telomeric repeats suddenly reverse direction - becoming (CCCTAA)3' instead of 5'(TTAGGG), and the second pre-telomeric section is also the reverse of the first telomeric section. This pattern is precisely as predicted by a telomere to telomere fusion of the chimpanzee (ancestor) 2p and 2q chromosomes, and in precisely the expected location. Note that the CCCTAA sequence is the reversed complement of TTAGGG (C pairs with G, and T pairs with A).Another piece of evidence is that if human Chromosome 2 had formed by chromosome fusion in an ancestor instead of being designed "as is", it should have evidence of 2 centromeres (the "pinched waist" in the picture above -- chromosomes have centromeres to aid in cell division). A "designed" chromosome would need only 1 centromere. An accidentally "merged" chromosome would show evidence of the 2 centromeres from the two chromosomes it merged from (one from each). And indeed, as documented in (Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M, Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet 1992 May;89(2):247-9), the functional centromere found on human Chromosome 2 lines up with the centromere of the chimp 2p chromosome, while there are non-functional remnants of the chimp 2q centromere at the expected location on the human chromosome.As an aside, the next time some creationist claims that there is "no evidence" for common ancestry or evolution, keep in mind that the sort of detailed "detective story" discussed above is repeated literally COUNTLESS times in the ordinary pursuit of scientific research and examination of biological and other types of evidence. Common ancestry and evolution is confirmed in bit and little ways over and over and over again. It's not just something that a couple of whacky anti-religionists dream up out of thin air and promulgate for no reason, as the creationists would have you believe.
For all practical purposes, the debate about common descent is *OVER*. Anyone who has really been reading the flood of DNA research papers which have come out in the past five years knows for a fact (yes, I said "fact") that common descent is now beyond any dispute. There are *volumes* of information of past life history in the gigabytes of DNA sequences of modern organisms, providing multiple, independent "tracers" of their evolutionary histories beyond any reasonable doubt. And anyone who disagrees with that is either unfamiliar with the studies, and/or doesn't understand them, and/or is lying about them.
Yes, that's a very strong statement. Yes, I stand by it 100%.
If you disagree, feel free to provide alternative explanations accounting for ALL the mountains of available evidence which explains *all* of the evidence (not just some small corner of it, *ALL* of it) in a way that a) makes actual sense, and b) somehow explains why all the evidence "just happens" to point strongly towards evolutionary processes if in fact they never actually happened.
Go for it.
And yes, I've had it up to *here* with arrogant know-nothings. If you've managed to live this long without learning a damned thing about science, but are willing to smugly pontificate on it and tell us all that 140+ years of biological research is not only all wrong, but somehow an empty shell to boot, then prepare to be hit over the head with a few hundred of the literally MILLIONS of research results of which you are so monumentally ignorant. As the immortal Red Skelton said in a classic sketch, "little do you know how little you know".
Wake up and learn something about the topic for a change, or for pete's sake, at least stop spreading your ignorance -- the world has more than enough of that commodity already.
eep!
reading that post is like witnessing a hurricane of anvils.
good post, bflr
Is this good for a juicy little grant for the seekers of PhD Welfare?

Disclaimer: Opinions posted on Free Republic are those of the individual posters and do not necessarily represent the opinion of Free Republic or its management. All materials posted herein are protected by copyright law and the exemption for fair use of copyrighted works.