To: lasereye; Big Red Badger; exDemMom; ifinnegan; Bob434; American in Israel
from the article:
"However, at least three of these papers described the amount of non-similar data that was thrown out.
When those missing data were included in the original numbers, an overall DNA similarity between humans and chimpanzees was only about 81 to 87%, depending on the paper! " Estimates of similarities in various species DNA were always dependent on the methodologies & logic used.
So, if we use one method to say that human DNA is 99.9% the same (understanding that means about 3 million differences!) and then a different method to say Neanderthals were 99% the same (30 million differences) and chimpanzees 98% (64 million differences), then such comparisons lose some meaning.
But obviously our author here, Mr. Thomkins, is using yet another methodology to arrive at a number which satisfies him, but seems to cast doubt on others.
I would question if Thomkins' methodology used on humans would still show us 99.9% the same genetically?
If not, then we might ask why Thomkins wants to show a larger number of human genetic differences than other methodologies produce?
151 posted on
06/03/2017 4:14:29 PM PDT by
BroJoeK
(a little historical perspective...)
To: BroJoeK
“I would question if Thomkins’ methodology used on humans would still show us 99.9% the same genetically?”
I think it would, actually. The issues of synteny and differencing chromosome numbers would not come in to play.
His method may or may not be more accurate than others. But it doesn’t change anything vis-a-vis common descent.
Phylogenetic results wouldn’t change, I don’t think. In other words, apes and humans would still be closest whether 98% or 85% etc... Down the line.
152 posted on
06/03/2017 4:31:31 PM PDT by
ifinnegan
(Democrats kill babies and harvest their organs to sell)
To: BroJoeK
153 posted on
06/03/2017 4:38:40 PM PDT by
Big Red Badger
(UNSCANABLE in an IDIOCRACY!)
To: BroJoeK
"Estimates of similarities in various species DNA were always dependent on the methodologies & logic used."
This is not clear. Are you referring to determining which sequences to compare? And this include or exclude (which seems to be this guy's point).
Blat and blast are the most used and best tools (their developers deserve the Nobel Prize in my opinion).
It is easy to compare splices coding sequences (eg mRNA or cDNA) from start to stop. And it's easy with genome data (ie chromosome sequence data) to compared start to stop including introns.
Both these provide a degree of identity or homology, the former will be much higher, yet the latter is the more true comparison, physically.
And then, at the genomic level, synteny plays a huge factor.
Look at this syntenic map of human and chimp.
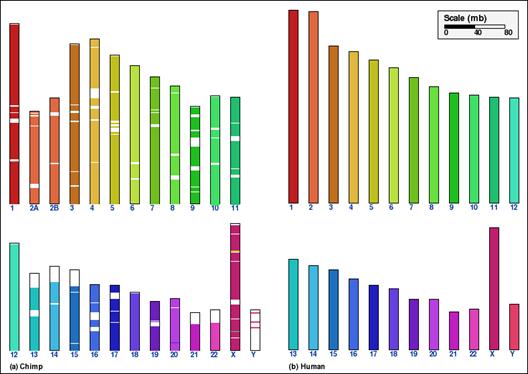
What does one do with the white regions?
And the degree of synteny is so high this isn't so hard compared to human mouse.
154 posted on
06/03/2017 4:47:31 PM PDT by
ifinnegan
(Democrats kill babies and harvest their organs to sell)
To: BroJoeK; exDemMom
And I post this one separately so it doesn’t get lost in the mix. Chromatin structure and regulation is essential for all aspects of biology. Ultimately this is what determines a species, eg why a human and ape genome can be so similar, especially in coding regions, but something causes one to be human, one ape.
The article below is representative of this type of work where the regulatory importance of the worst of the worst “junk DNA” is being elucidated and is crucial at a fundamental level.
—/
Chromosome Res. 2017 Mar;25(1):77-87. doi: 10.1007/s10577-016-9547-3. Epub 2017 Jan 11.
The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals.
Nishibuchi G1, Déjardin J2.
Author information
1
Biology of Repetitive Sequences, CNRS UPR1142, 141 rue de la Cardonille, 34000, Montpellier, France.
2
Biology of Repetitive Sequences, CNRS UPR1142, 141 rue de la Cardonille, 34000, Montpellier, France. jerome.dejardin@igh.cnrs.fr.
Abstract
Constitutive heterochromatin is composed mainly of repetitive elements and represents the typical inert chromatin structure in eukaryotic cells. Approximately half of the mammalian genome is made of repeat sequences, such as satellite DNA, telomeric DNA, and transposable elements. As essential genes are not present in these regions, most of these repeat sequences were considered as junk DNA in the past. However, it is now clear that these regions are essential for chromosome stability and the silencing of neighboring genes. Genetic and biochemical studies have revealed that histone methylation at H3K9 and its recognition by heterochromatin protein 1 represent the fundamental mechanism by which heterochromatin forms. Although this molecular mechanism is highly conserved from yeast to human cells, its detailed epigenetic regulation is more complex and dynamic for each distinct constitutive heterochromatin structure in higher eukaryotes. It can also vary according to the developmental stage. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis is a powerful tool to investigate the epigenetic regulation of eukaryote genomes, but non-unique reads are usually discarded during standard ChIP-seq data alignment to reference genome databases. Therefore, specific methods to obtain global epigenetic information concerning repetitive elements are needed. In this review, we focus on such approaches and we summarize the latest molecular models for distinct constitutive heterochromatin types in mammals.
KEYWORDS:
Constitutive heterochromatin; endogenous retrovirus; interspersed repetitive element; pericentromere; retrotransposon; tandem repeat; telomere
PMID: 28078514 DOI: 10.1007/s10577-016-9547-3
195 posted on
06/04/2017 11:54:22 AM PDT by
ifinnegan
(Democrats kill babies and harvest their organs to sell)
FreeRepublic.com is powered by software copyright 2000-2008 John Robinson
