He undoubtedly had a lot more than *this* author...
With an ostrich mindset that tries to ignore reality, pseudo-scientists continue in the vain hope that if they shout loud and long enough they can perpetuate the fairy story and bad science that is evolution.
Blah, blah, blah. Sorry, but evolutionary biology is an extremely well-established science, grounded in a *vast* amount of evidence and research along multiple independently cross-confirming lines, which has passed a mind-boggling number of various validation checks and attempts at falsification. Deal with it. No amount of empty blustering, no amount of childishly labeling it "fairy story", "bad science", or "pseudo-science", etc., is going to change that, nor is dishonestly pretending that evolutionary biology is only supported by "shouting loudly and longly", as if it doesn't have an *enormous* body of evidence and research to bring to bear.
You don't have to be a religious fundamentalist to question evolution theory
...but it sure helps. The great majority of people strenuously "questioning" evolution in essays, editorials, school boards, etc., do so primarily due to religious objections, and it's disingenuous of the author to try to pretend otherwise.
- you just have to have an open and enquiring mind and not be afraid of challenging dogma.
...and have deep-seated misunderstandings of the sort that makes you mistake established science for "dogma", etc.
Most people who actually have "open and enquiring minds" learn enough about the biology and evidence that they, like the vast majority (99+%) of biologists, come to understand that the evidence and research does, indeed, make an overwhelming and consistent case for the reality of evolution, and accept the validity of this field of science.
It's only the people whose ideas of "open and enquring minds" means reading as many anti-evolution pamphlets as they can in order to have more ammo (no matter how poor) to attack evolutionary biology -- WITHOUT spending much time, if any, actually wading through primary research papers (or even a few good college-level biology textbooks) in order to actually *understand* anything about the subject they're attempting to attack.
But you must be able to discern and dodge the effusion of evolutionary landmines that are bluster and non sequiturs.
Funny, the author strives hard to imply that there's an "effusion" of fallacies in evolutionary biology, but he doesn't actually *identify* any... Not very honest of him, is it? Furthermore, he makes quite a lot of "bluster and non sequiturs" himself, as we shall see below.
No one denies the reality of variation and natural selection.
Actually, many anti-evolutionists deny even that, but no matter.
For example, chihuahuas and Great Danes can be derived from a wolf by selective breeding. Therefore, a chihuahua is a wolf, in the same way that people of short stature and small brain capacity are fully human beings.
No, sorry, chihuahuas are no longer wolves, they're domestic dogs. Nice try. No one in their right mind, encountering a chihuahua for the first time, would conclude, "oh look, it's a miniature wolf!" The author dishonestly attempts to hand-wave away significant evolutionary change by providing an example, then simply declaring (with no evidence or justification offered) that it isn't "really" a change at all. Bizarre.
However, there is no evidence (fossil, anatomical, biochemical or genetic) that any creature did give rise, or could have given rise, to a different creature.
This is just such an incredibly transparently false statement that one has to wonder whether the author is truly this vastly ignorant of his subject, or merely a bald-faced liar. I could take the author into any decent research library and literally *bury* him using just a *small* portion of the evidence (fossil, anatomical, biochemical *and* genetic) for macroevolution.
For just the tip of the iceberg, here's a table of contents of some web pages which outline over TWO DOZEN independent lines of evidence which cross-confirm each other and establish the validity of macroevolution beyond any reasonable doubt:
[From: http://www.talkorigins.org/faqs/comdesc/]And don't make the mistake of thinking that the short descriptions on these handful of web pages encompasses the totality of the evidence -- it's just an overview. Go follow any of the hundreds of citations provided, and you'll find your entry to the primary research literature, containing *vast* amounts of rigorous research and evidence, and a web of citations leading to literally *millions* of other papers which embody the evidence for evolution.
29+ Evidences for Macroevolution
The Scientific Case for Common Descent
Version 2.85
Copyright © 1999-2004 by Douglas Theobald, Ph.D.[Last Update: April 15, 2005]Permission is granted to copy and print these pages in total for non-profit personal, educational, research, or critical purposes.
Introduction
volution, the overarching concept that unifies the biological sciences, in fact embraces a plurality of theories and hypotheses. In evolutionary debates one is apt to hear evolution roughly parceled between the terms "microevolution" and "macroevolution". Microevolution, or change beneath the species level, may be thought of as relatively small scale change in the functional and genetic constituencies of populations of organisms. That this occurs and has been observed is generally undisputed by critics of evolution. What is vigorously challenged, however, is macroevolution. Macroevolution is evolution on the "grand scale" resulting in the origin of higher taxa. In evolutionary theory it thus entails common ancestry, descent with modification, speciation, the genealogical relatedness of all life, transformation of species, and large scale functional and structural changes of populations through time, all at or above the species level (Freeman and Herron 2004; Futuyma 1998; Ridley 1993).
Common descent is a general descriptive theory that concerns the genetic origins of living organisms (though not the ultimate origin of life). The theory specifically postulates that all of the earth's known biota are genealogically related, much in the same way that siblings or cousins are related to one another. Thus, macroevolutionary history and processes necessarily entail the transformation of one species into another and, consequently, the origin of higher taxa. Because it is so well supported scientifically, common descent is often called the "fact of evolution" by biologists. For these reasons, proponents of special creation are especially hostile to the macroevolutionary foundation of the biological sciences.
This article directly addresses the scientific evidence in favor of common descent and macroevolution. This article is specifically intended for those who are scientifically minded but, for one reason or another, have come to believe that macroevolutionary theory explains little, makes few or no testable predictions, is unfalsifiable, or has not been scientifically demonstrated.
Outline
- Universal Common Descent Defined
- Evidence for Common Descent is Independent of Mechanism
- What Counts as Scientific Evidence
- Other Explanations for the Biology
- How to Cite This Document
The kind of incredible ignorance which leads an embarrassingly large number of anti-evolutionists to mouth such shockingly stupid statements as "there is no evidence for evolution, blah blah blah" is just mind-boggling. And make no mistake -- millions of Americans *are* familiar with the evidence for evolution, at least to some degree, and when so-called conservatives make such blindingly moronic comments as "there's no evidence", it's *entirely* obvious to informed Americans that the speaker is a dolt, and this does *not* reflect well on conservatism in general. I personally know *dozens* of people who would otherwise be predisposed to consider themselves conservatives or vote Republican, who are utterly repulsed from the idea because they've seen too many conservatives bleat idiocy like this, and it scares them away -- in exactly the same reason that lots of liberal-leannig folks have run screaming from the Democrats because of the nutball rantings of some of the more fringe liberals.
In addition, by their absence in the fossil record for (supposed) millions of years along with the fact of their existence during the same time period, many animals such as the coelacanth demonstrate the principle that all creatures could have lived contemporaneously in the past.
Utter nonsense:
Creationist Claim CB930.1: The coelacanthIn short, the reason that the more recent descendants of the ancient coelacanth haven't left fossils in areas where we could find them are well understood by biologists, and those reasons do *not* apply to most of the vast numbers of species which the author would like to pretend might be missing from the early fossil record for the same reason. Nice try, but he's just engaging in ignorant handwaving. His excuses just don't hold water.
No evidence supports the notion that birds evolved from dinosaurs
Again, the amount of ignorance in this false claim is just boggling. There is a vast amount of evidence "supporting the notion", including for example a very well populated and cladistically consistent sequence of transitional fossils (again from a prior post of mine):
The biochemical and genetic evidence for common ancestry between reptiles and birds is even more overwhelming. The author sort of "forgot" to mention that too.Theropod dinosaur to bird evolutionary transition:
The cladogram for the evolution of flight looks like this:
(Note -- each name along the top is a known transitional fossil; and those aren't all that have been discovered.) Here's a more detailed look at the middle section:
Fossils discovered in the past ten years in China have answered most of the "which came first" questions about the evolution of birds from dinosaurs.
We now know that downy feathers came first, as seen in this fossil of Sinosauropteryx:
That's a close-up of downy plumage along the backbone. Here's a shot of an entire fossil
Sinosauropteryx was reptilian in every way, not counting the feathers. It had short forelimbs, and the feathers were all the same size. Presumably, the downy feathers evolved from scales driven by a need for bodily insulation.
Next came Protarchaeopteryx:
It had long arms, broad "hands", and long claws:
Apparently this species was driven by selection to develop more efficient limbs for grasping prey. One of the interesting things about this species is that the structure of the forelimb has been refined to be quite efficient at sweeping out quickly to grab prey, snap the hands together, then draw them back towards the body (mouth?). The specific structures in question are the semilunate carpal (a wrist bone), that moves with the hand in a broad, flat, 190 degree arc, heavy chest muscles, bones of the arm which link together with the wrist so as to force the grasping hands to spread out toward the prey during the forestroke and fold in on the prey during the upstroke. Not only is this a marvelously efficient prey-grabbing mechanism, but the same mechanism is at the root of the wing flight-stroke of modern birds. Evolution often ends up developing a structure to serve one need, then finds it suitable for adaptation to another. Here, a prey-grasping motion similar in concept to the strike of a praying mantis in a reptile becomes suitable for modifying into a flapping flight motion.
Additionally, the feathers on the hands and tail have elongated, becoming better suited for helping to sweep prey into the hands.
Next is Caudipteryx:
This species had hand and tail feathers even more developed than the previous species, and longer feathers, more like that of modern birds:
However, it is clear that this was still not a free-flying animal yet, because the forelimbs were too short and the feathers not long enough to support its weight, and the feathers were symmetrical (equal sized "fins" on each side of the central quill). It also had very reduced teeth compared to earlier specimens and a stubby beak:
But the elongation of the feathers indicates some aerodynamic purpose, presumably gliding after leaping (or falling) from trees which it had climbed with its clawed limbs, in the manner of a flying squirrel. Feathers which were developed "for" heat retention and then pressed into service to help scoop prey were now "found" to be useful for breaking falls or gliding to cover distance (or swooping down on prey?).
Next is Sinornithosaurus:
Similar to the preceding species, except that the pubis bone has now shifted to point to the back instead of the front, a key feature in modern birds (when compared to the forward-facing publis bone in reptiles). Here are some of the forearm feathers in detail:
Long feathers in detail:
Artists' reconstruction:
Next is Archaeopteryx:
The transition to flight is now well underway. Archaeopteryx has the reversed hallux (thumb) characteristic of modern birds, and fully developed feathers of the type used for flight (long, aligned with each other, and assymetrical indicating that the feathers have been refined to function aerodynamically). The feathers and limbs are easily long enough to support the weight of this species in flight. However, it lacks some structures which would make endurance flying more practical (such as a keeled sternum for efficient anchoring of the pectoral muscles which power the downstroke) and fused chest vertebrae. Archaeopteryx also retains a number of clearly reptilian features still, including a clawed "hand" emerging from the wings, small reptilian teeth, and a long bony tail. After the previous species' gliding abilities gave it an advantage, evolution would have strongly selected for more improvements in "flying" ability, pushing the species towards something more resembling sustained powered flight.
Next is Confuciusornis:
This species had a nearly modern flight apparatus. It also displays transitional traits between a reptilian grasping "hand" and a fully formed wing as in modern birds -- the outer two digits (the earlier species had three-fingered "hands") in Confuciusornis are still free, but the center digit has now formed flat, broad bones as seen in the wings of modern birds.
Additionally, the foot is now well on its way towards being a perching foot as in modern birds:
It also has a keeled sternum better suited for long flight, and a reduced number of vertebrae in the tail, on its way towards becoming the truncated tail of modern birds (which while prominent, is a small flap of muscle made to look large only because of the long feathers attached).
From this species it's only a small number of minor changes to finish the transition into the modern bird family.
(Hey, who said there are no transitional fossils? Oh, right, a lot of dishonest creationists. And there are a lot more than this, I've just posted some of the more significant milestones.)
There's been a very recent fossil find along this same lineage, too new for me to have found any online images to include in this article. And analysis is still underway to determine exactly where it fits into the above lineage. But it has well-formed feathers, which extend out from both the "arms" and the legs. Although it wasn't advanced enough to fully fly, the balanced feathering on the front and back would have made it ideally suited for gliding like a flying squirrel, and it may be another link between the stage where feathers had not yet been pressed into service as aerodynamic aids, and the time when they began to be used more and more to catch the air and developing towards a "forelimbs as wings" specialization.
So in short, to answer your question about how flight could have developed in birds, the progression is most likely some minor refinement on the following:
1. Scales modified into downy feathers for heat retention.
2. Downy feathers modified into "straight" feathers for better heat retention (modern birds still use their body "contour feathers" in this fashion).
3. Straight feathers modified into a "grasping basket" on the hands (with an accompanying increase in reach for the same purpose).
4. Long limbs with long feathers refined to better survive falls to the ground.
5. "Parachute" feathers refined for better control, leading to gliding.
6. Gliding refined into better controlled, longer gliding.
7. Long gliding refined into short powered "hops".
8. Short powered flight refined into longer powered flight.
9. Longer powered flight refined into long-distance flying.Note that in each stage, the current configuration has already set the stage for natural selection to "prefer" individuals which better meet the requirements of the next stage. Evolution most often works like this; by taking some pre-existing ability or structure, and finding a better use for it or a better way to make it perform its current use.
nor that whales evolved from terrestrial quadrupeds
Yeah, right, sure. This guy's really on a role when he's spewing the falsehoods, isn't he? So there's "no evidence supporting" that whales evolved from terrestrial quadrupeds, eh? Little does he know how little he knows:
Whale Evolution
The transitional fossils in the evolutionary origin of whales is especially striking. The following is an excerpt from The Origin of Whales and the Power of Independent Evidence . This excerpt is excellent all by itself, but one should really read the entire essay in order to get the "big picture" of whale evolution:The evidenceThe paleontological (i.e. fossil) evidence for evolutionary transitions is overwhelming to anyone who has actually examined the evidence with an open mind. However, a stubborn person attempting to deny the obvious can rationalize it away by refusing to see the clear sequences of morphological change, and insisting that one can't "prove" that the various fossil specimens are "really" necessarily related. That excuse crumbles when one compares the fossil evidence to the *many* other independent lines of evidence which confirm the fossil evidence. For example, concerning whale evolution:
The evidence that whales descended from terrestrial mammals is here divided into nine independent parts: paleontological, morphological, molecular biological, vestigial, embryological, geochemical, paleoenvironmental, paleobiogeographical, and chronological. Although my summary of the evidence is not exhaustive, it shows that the current view of whale evolution is supported by scientific research in several distinct disciplines.
1. Paleontological evidence
The paleontological evidence comes from studying the fossil sequence from terrestrial mammals through more and more whale-like forms until the appearance of modern whales. Although the early whales (Archaeocetes) exhibit greater diversity than I have space to discuss here, the examples in this section represent the trends that we see in this taxon. Although there are two modern suborders of whales (Odontocetes and Mysticetes), this discussion will focus on the origin of the whales as an order of mammals, and set aside the issues related to the diversification into suborders.
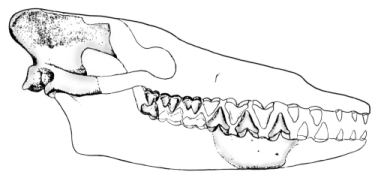
Sinonyx
We start with Sinonyx, a wolf-sized mesonychid (a primitive ungulate from the order Condylarthra, which gave rise to artiodactyls, perissodactyls, proboscideans, and so on) from the late Paleocene, about 60 million years ago. The characters that link Sinonyx to the whales, thus indicating that they are relatives, include an elongated muzzle, an enlarged jugular foramen, and a short basicranium (Zhou and others 1995). The tooth count was the primitive mammalian number (44); the teeth were differentiated as are the heterodont teeth of today's mammals. The molars were very narrow shearing teeth, especially in the lower jaw, but possessed multiple cusps. The elongation of the muzzle is often associated with hunting fish - all fish-hunting whales, as well as dolphins, have elongated muzzles. These features were atypical of mesonychids, indicating that Sinonyx was already developing the adaptations that later became the basis of the whales' specialized way of life.
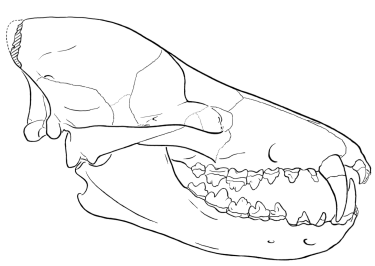
Pakicetus
The next fossil in the sequence, Pakicetus, is the oldest cetacean, and the first known archaeocete. It is from the early Eocene of Pakistan, about 52 million years ago (Gingerich and others 1983). Although it is known only from fragmentary skull remains, those remains are very diagnostic, and they are definitely intermediate between Sinonyxand later whales. This is especially the case for the teeth. The upper and lower molars, which have multiple cusps, are still similar to those of Sinonyx, but the premolars have become simple triangular teeth composed of a single cusp serrated on its front and back edges. The teeth of later whales show even more simplification into simple serrated triangles, like those of carnivorous sharks, indicating that Pakicetus's teeth were adapted to hunting fish.
Gingrich and others (1983) published this reconstruction of the skull of
Pakicetus inachus (redrawn for RNCSE by Janet Dreyer).
A well-preserved cranium shows that Pakicetus was definitely a cetacean with a narrow braincase, a high, narrow sagittal crest, and prominent lambdoidal crests. Gingerich and others (1983) reconstructed a composite skull that was about 35 centimeters long. Pakicetus did not hear well underwater. Its skull had neither dense tympanic bullae nor sinuses isolating the left auditory area from the right one - an adaptation of later whales that allows directional hearing under water and prevents transmission of sounds through the skull (Gingerich and others 1983). All living whales have foam-filled sinuses along with dense tympanic bullae that create an impedance contrast so they can separate sounds arriving from different directions. There is also no evidence in Pakicetus of vascularization of the middle ear, which is necessary to regulate the pressure within the middle ear during diving (Gingerich and others 1983). Therefore, Pakicetus was probably incapable of achieving dives of any significant depth. This paleontological assessment of the ecological niche of Pakicetus is entirely consistent with the geochemical and paleoenvironmental evidence. When it came to hearing, Pakicetus was more terrestrial than aquatic, but the shape of its skull was definitely cetacean, and its teeth were between the ancestral and modern states.
Zhou and others (1995) published this reconstruction of the skull of
Sinonyx jiashanensis (redrawn for RNCSE by Janet Dreyer).
Ambulocetus
In the same area that Pakicetus was found, but in sediments about 120 meters higher, Thewissen and colleagues (1994) discovered Ambulocetus natans, "the walking whale that swims", in 1992. Dating from the early to middle Eocene, about 50 million years ago, Ambulocetus is a truly amazing fossil. It was clearly a cetacean, but it also had functional legs and a skeleton that still allowed some degree of terrestrial walking. The conclusion that Ambulocetus could walk by using the hind limbs is supported by its having a large, stout femur. However, because the femur did not have the requisite large attachment points for walking muscles, it could not have been a very efficient walker. Probably it could walk only in the way that modern sea lions can walk - by rotating the hind feet forward and waddling along the ground with the assistance of their forefeet and spinal flexion. When walking, its huge front feet must have pointed laterally to a fair degree since, if they had pointed forward, they would have interfered with each other.
The forelimbs were also intermediate in both structure and function. The ulna and the radius were strong and capable of carrying the weight of the animal on land. The strong elbow was strong but it was inclined rearward, making possible rearward thrusts of the forearm for swimming. However, the wrists, unlike those of modern whales, were flexible.
It is obvious from the anatomy of the spinal column that Ambulocetus must have swum with its spine swaying up and down, propelled by its back feet, oriented to the rear. As with other aquatic mammals using this method of swimming, the back feet were quite large. Unusually, the toes of the back feet terminated in hooves, thus advertising the ungulate ancestry of the animal. The only tail vertebra found is long, making it likely that the tail was also long. The cervical vertebrae were relatively long, compared to those of modern whales; Ambulocetus must have had a flexible neck.
Ambulocetus's skull was quite cetacean (Novacek 1994). It had a long muzzle, teeth that were very similar to later archaeocetes, a reduced zygomatic arch, and a tympanic bulla (which supports the eardrum) that was poorly attached to the skull. Although Ambulocetus apparently lacked a blowhole, the other skull features qualify Ambulocetus as a cetacean. The post-cranial features are clearly in transitional adaptation to the aquatic environment. Thus Ambulocetus is best described as an amphibious, sea-lion-sized fish-eater that was not yet totally disconnected from the terrestrial life of its ancestors.
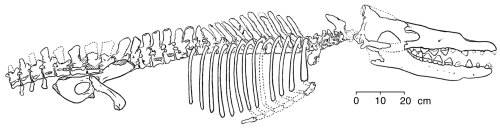
Rodhocetus
In the middle Eocene (46-7 million years ago) Rodhocetus took all of these changes even further, yet still retained a number of primitive terrestrial features (Gingerich and others 1994). It is the earliest archaeocete of which all of the thoracic, lumbar, and sacral vertebrae have been preserved. The lumbar vertebrae had higher neural spines than in earlier whales. The size of these extensions on the top of the vertebrae where muscles are attached indicate that Rodhocetus had developed a powerful tail for swimming.
Gingrich and others (1994) published this reconstruction of the skeleton of
Rodhocetus kasrani (redrawn for RNCSE by Janet Dreyer).
Elsewhere along the spine, the four large sacral vertebrae were unfused. This gave the spine more flexibility and allowed a more powerful thrust while swimming. It is also likely that Rodhocetus had a tail fluke, although such a feature is not preserved in the known fossils: it possessed features - shortened cervical vertebrae, heavy and robust proximal tail vertebrae, and large dorsal spines on the lumbar vertebrae for large tail and other axial muscle attachments - that are associated in modern whales with the development and use of tail flukes. All in all, Rodhocetus must have been a very good tail-swimmer, and it is the earliest fossil whale committed to this manner of swimming.
The pelvis of Rodhocetus was smaller than that of its predecessors, but it was still connected to the sacral vertebrae, meaning that Rodhocetus could still walk on land to some degree. However, the ilium of the pelvis was short compared to that of the mesonychids, making for a less powerful muscular thrust from the hip during walking, and the femur was about 1/3 shorter than Ambulocetus’s, so Rodhocetus probably could not get around as well on land as its predecessors (Gingerich and others 1994).
Rodhocetus's skull was rather large compared to the rest of the skeleton. The premaxillae and dentaries had extended forward even more than its predecessors’, elongating the skull and making it even more cetacean. The molars have higher crowns than in earlier whales and are greatly simplified. The lower molars are higher than they are wide. There is a reduced differentiation among the teeth. For the first time, the nostrils have moved back along the snout and are located above the canine teeth, showing blowhole evolution. The auditory bullae are large and made of dense bone (characteristics unique to cetaceans), but they apparently did not contain the sinuses typical of later whales, making it questionable whether Rodhocetus possessed directional hearing underwater.
Overall, Rodhocetus showed improvements over earlier whales by virtue of its deep, slim thorax, longer head, greater vertebral flexibility, and expanded tail-related musculature. The increase in flexibility and strength in the back and tail with the accompanying decrease in the strength and size of the limbs indicated that it was a good tail-swimmer with a reduced ability to walk on land.
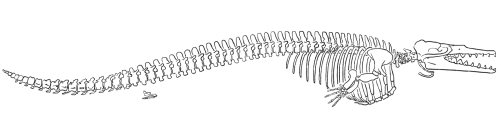
Basilosaurus
The particularly well-known fossil whale Basilosaurus represents the next evolutionary grade in whale evolution (Gingerich 1994). It lived during the late Eocene and latest part of the middle Eocene (35-45 million years ago). Basilosaurus was a long, thin, serpentine animal that was originally thought to have been the remains of a sea serpent (hence it is name, which actually means "king lizard"). Its extreme body length (about 15 meters) appears to be due to a feature unique among whales; its 67 vertebrae are so long compared to other whales of the time and to modern whales that it probably represents a specialization that sets it apart from the lineage that gave rise to modern whales.
What makes Basilosaurus a particularly interesting whale, however, is the distinctive anatomy of its hind limbs (Gingerich and others 1990). It had a nearly complete pelvic girdle and set of hindlimb bones. The limbs were too small for effective propulsion, less than 60 cm long on this 15-meter-long animal, and the pelvic girdle was completely isolated from the spine so that weight-bearing was impossible. Reconstructions of the animal have placed its legs external to the body - a configuration that would represent an important intermediate form in whale evolution.
Although no tail fluke has ever been found (since tail flukes contain no bones and are unlikely to fossilize), Gingerich and others (1990) noted that Basilosaurus's vertebral column shares characteristics of whales that do have tail flukes. The tail and cervical vertebrae are shorter than those of the thoracic and lumbar regions, and Gingerich and others (1990) take these vertebral proportions as evidence that Basilosaurus probably also had a tail fluke.
Further evidence that Basilosaurus spent most of its time in the water comes from another important change in the skull. This animal had a large single nostril that had migrated a short distance back to a point corresponding to the back third of the dental array. The movement from the forward extreme of the snout to the a position nearer the top of the head is characteristic of only those mammals that live in marine or aquatic environments.
Dorudon
Dorudon was a contemporary of Basilosaurus in the late Eocene (about 40 million years ago) and probably represents the group most likely to be ancestral to modern whales (Gingerich 1994). Dorudon lacked the elongated vertebrae of Basilosaurus and was much smaller (about 4-5 meters in length). Dorudon’s dentition was similar to Basilosaurus’s; its cranium, compared to the skulls of Basilosaurus and the previous whales, was somewhat vaulted (Kellogg 1936). Dorudon also did not yet have the skull anatomy that indicates the presence of the apparatus necessary for echolocation (Barnes 1984).
Gingrich and Uhen (1996) published this reconstruction of the skeleton of
Dorudon atrox (redrawn for RNCSE by Janet Dreyer).
Basilosaurus and Dorudon were fully aquatic whales (like Basilosaurus, Dorudon had very small hind limbs that may have projected slightly beyond the body wall). They were no longer tied to the land; in fact, they would not have been able to move around on land at all. Their size and their lack of limbs that could support their weight made them obligate aquatic mammals, a trend that is elaborated and reinforced by subsequent whale taxa.
Clearly, even if we look only at the paleontological evidence, the creationist claim of "No fossil intermediates!" is wrong. In fact, in the case of whales, we have several, beautifully arranged in morphological and chronological order.
In summarizing the paleontological evidence, we have noted the consistent changes that indicate a series of adaptations from more terrestrial to more aquatic environments as we move from the most ancestral to the most recent species. These changes affect the shape of the skull, the shape of the teeth, the position of the nostrils, the size and structure of both the forelimbs and the hindlimbs, the size and shape of the tail, and the structure of the middle ear as it relates to directional hearing underwater and diving. The paleontological evidence records a history of increasing adaptation to life in the water - not just to any way of life in the water, but to life as lived by contemporary whales.
Evolution of whales from terrestrial mammals
(From Plagiarized Errors and Molecular Genetics)That's just a quick layman-level overview of *one* of the many ways that whale evolution has been verified. For more technical examinations along several independent lines of evidence, see for example:.
A particularly impressive example of shared retroposons has recently been reported linking cetaceans (whales, dolphins and porpoises) to ruminants and hippopotamuses, and it is instructive to consider this example in some detail. Cetaceans are sea-living animals that bear important similarities to land-living mammals; in particular, the females have mammary glands and nurse their young. Scientists studying mammalian anatomy and physiology have demonstrated greatest similarities between cetaceans and the mammalian group known as artiodactyls (even-toed ungulates) including cows, sheep, camels and pigs. These observations have led to the evolutionist view that whales evolved from a four-legged artiodactyl ancestor that lived on land. Creationists have capitalized on the obvious differences between the familiar artiodactyls and whales, and have ridiculed the idea that whales could have had four-legged land-living ancestors. Creationists who claim that cetaceans did not arise from four-legged land mammals must ignore or somehow dismiss the fossil evidence of apparent whale ancestors looking exactly like one would predict for transitional species between land mammals and whales--with diminutive legs and with ear structures intermediate between those of modern artiodactyls and cetaceans (Nature 368:844,1994; Science 263: 210, 1994). (A discussion of fossil ancestral whale species with references may be found at http://www.talkorigins.org/faqs/faq-transitional/part2b.html#ceta) Creationists must also ignore or dismiss the evidence showing the great similarity between cetacean and artiodactyl gene sequences (Molecular Biology & Evolution 11:357, 1994; ibid 13: 954, 1996; Gatesy et al, Systematic Biology 48:6, 1999).
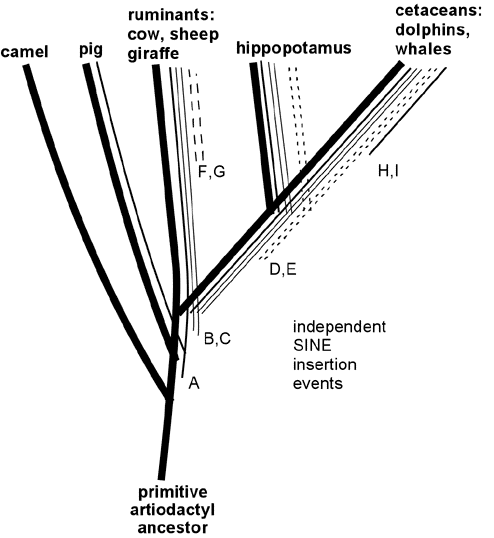
Recently retroposon evidence has solidified the evolutionary relationship between whales and artiodactyls. Shimamura et al. (Nature 388:666, 1997; Mol Biol Evol 16: 1046, 1999; see also Lum et al., Mol Biol Evol 17:1417, 2000; Nikaido and Okada, Mamm Genome 11:1123, 2000) studied SINE sequences that are highly reduplicated in the DNA of all cetacean species examined. These SINES were also found to be present in the DNA of ruminants (including cows and sheep) but not in DNA of camels and pigs or more distantly related mammals such as horse, elephant, cat, human or kangaroo. These SINES apparently originated in a specific branch of ancestral artiodactyls after this branch diverged from camels, pigs and other mammals, but before the divergence of the lines leading to modern cetaceans, hippopotamus and ruminants. (See Figure 5.) In support of this scenario, Shimamura et al. identified two specific insertions of these SINES in whale DNA (insertions B and C in Figure 5) and showed that in DNA of hippopotamus, cow and sheep these same two sites contained the SINES; but in camel and pig DNA the same sites were "empty" of insertions. More recently, hippopotamus has been identified as the closest living terrestrial relative of cetaceans since hippos and whales share retroposon insertions (illustrated by D and E in Figure 5) that are not found in any other artiodactyls (Nikaido et al, PNAS 96:10261, 1999). The close hippo-whale relationship is consistent with previously reported sequence similarity comparisons (Gatesy, Mol Biol Evol 14:537, 1997) and with recent fossil finds (Gingerich et al., Science 293:2239, 2001; Thewissen et al., Nature 413:277, 2001) that resolve earlier paleontological conflicts with the close whale-hippo relationship. (Some readers have wondered: if ruminants are more closely related to whales than to pigs and camels, why are ruminants anatomically more similar to pigs and camels than they are to whales? Apparently this results from the fact that ruminants, pigs and camels changed relatively little since their last common ancestor, while the cetacean lineage changed dramatically in adapting to an aquatic lifestyle, thereby obliterating many of the features -- like hooves, fur and hind legs -- that are shared between its close ruminant relatives and the more distantly related pigs and camels. This scenario illustrates the fact that the rapid evolutionary development of adaptations to a new niche can occur through key functional mutations, leaving the bulk of the DNA relatively unchanged. The particularly close relationship between whales and hippos is consistent with several shared adaptations to aquatic life, including use of underwater vocalizations for communication and the absence of hair and sebaceous glands.) Thus, retroposon evidence strongly supports the derivation of whales from a common ancestor of hippopotamus and ruminants, consistent with the evolutionary interpretation of fossils and overall DNA sequence similarities. Indeed, the logic of the evidence from shared SINEs is so powerful that SINEs may be the best available characters for deducing species relatedness (Shedlock and Okada, Bioessays 22:148, 2000), even if they are not perfect (Myamoto, Curr. Biology 9:R816, 1999).
Figure 5. Specific SINE insertions can act as "tracers" that illuminate phylogenetic relationships. This figure summarizes some of the data on SINEs found in living artiodactyls and shows how the shared insertions can be interpreted in relation to evolutionary branching. A specific SINE insertion event ("A" in the Figure) apparently occurred in a primitive common ancestor of pigs, ruminants, hippopotamus and cetaceans, since this insertion is present in these modern descendants of that common ancestor; but it is absent in camels, which split off from the other species before this SINE inserted. More recent insertions B and C are present only in ruminants, hippopotamus and cetaceans. Insertions D and E are shared only by hippopotamus and cetaceans, thereby identifying hippopotamus as the closest living relative of cetaceans (at least among the species examined in these studies). SINE insertions F and G occurred in the ruminant lineage after it diverged from the other species; and insertions H and I occurred after divergence of the cetacean lineage.
SINE Evolution, Missing Data, and the Origin of WhalesAnd much, much more.Evidence from Milk Casein Genes that Cetaceans are Close Relatives of Hippopotamid Artiodactyls
Analyses of mitochondrial genomes strongly support a hippopotamus±whale clade
A new Eocene archaeocete (Mammalia, Cetacea) from India and the time of origin of whales
Mysticete (Baleen Whale) Relationships Based upon the Sequence of the Common Cetacean DNA Satellite1
Eocene evolution of whale hearing
Novel Phylogeny of Whales Revisited but Not Revised
New Morphological Evidence for the Phylogeny of Artiodactyla, Cetacea, and Mesonychidae
nor that the human knee joint evolved from a fish pelvic fin.
Wrong again! There's a very decent set of transitional fossils from ray-finned fish to amphibians. This alone establishes the evolution from "fish pelvic fin" to the standard terrestrial vertebrate leg. And if a clear fossil sequence isn't enough for you (creationists in the back scream, as always, "that don't prove nothin'!") there's corresponding corroboratnig biochemical/genetic evidence of fish/amphibian common ancestry. The same (fossil/biochemical/genetic cross-linked evidence) for amphibian->reptile and reptile->mammal common ancestry and evolutionary transition, and from there the transition from protomammal legs to "human knee joint" is mapped out in pretty good detail as well.
Laying out all of the available evidence on just this *one* subject would fill several large encyclopedia-sized volumes, so when the author claims that there is "no evidence" supporting this, ponder just how big of an idiot or a liar he would have to be to make such a whopper with a straight face.
And the critically-positioned amino acids at the active sites within enzymes and structural proteins show that the origination of complex proteins by step-wise modifications of supposed ancestral peptides is impossible.
No, it doesn't. Biochemical evolution has been studied in incredible detail by tens of thousands of researchers for many decades now, and found to be quite mundane and very "possible" indeed, to the point where it's commonly harnessed in order to produce novel new pharmaceuticals, as well as directly observed in biology labs all around the world on a daily basis. The author is, in a word, an idiot. Or a liar. You decide which.
In other words, birds have always been birds, whales have always been whales, apes did not evolve into humans, and humans have always been humans.
In other words, the author's claims have all fallen flat on their faces because they contradict reality, and his conclusion is therefore not only unsupported, but in fact is contradicted by the actual evidence he pretends doesn't exist but which actually exists in vast abundance.
But you might protest that it has been proved that we evolved from apes.
No I wouldn't, because science does not deal in "proofs". It's hilarious that the author is so ignorant of science as to think that it does, especially when he started out this rant by snottily declaring that someone *else* "appears to have little understanding of biology". Psychological projection?
I would, however, point out that the common ancestry of humans and modern apes has been established by vast amounts of evidence beyond any reasonable doubt (to anyone who actually goes and *looks* at all the evidence, that is, which leaves out most anti-evolutionists), because it has. The biochemical/genetic evidence *alone* demonstrated this so conclusively in the past couple of decades that to anyone with an open mind, the issue is *over*. The biochemical/genetic evidence has so firmly established the fact that the evolutionists are right, and the anti-evolutionists are wrong, that the only holdouts are people as ignorant of the evidence as this author is ("evidence, what evidence, there ain't no stinkin' evidence!"), or who are so wedded to their preconceptions that they can rationalize a cheap excuse to ignore *any* amount of evidence, instead of being intellectually honest enough to follow it wherever it actually leads.
For just some examples, some mere tips of the iceberg of the vast amount of evidence establishing human/ape common ancestry as firmly as the DNA and other evidence placing O.J. Simpson at the murder scene (again from prior posts of mine):
And:Background: Retroviruses reproduce by entering a cell of a host (like, say, a human), then embedding their own viral DNA into the cell's own DNA, which has the effect of adding a "recipe" for manufacturing more viruses to the cell's "instruction book". The cell then follows those instructions because it has no reason (or way) to "mistrust" the DNA instructions it contains. So the virus has converted the cell into a virus factory, and the new viruses leave the cell, and go find more cells to infect, etc.
However, every once in a while a virus's invasion plans don't function exactly as they should, and the virus's DNA (or portions of it) gets embedded into the cell's DNA in a "broken" manner. It's stuck into there, becoming part of the cell's DNA, but it's unable to produce new viruses. So there it remains, embedded in the DNA. If this happens in a regular body cell, it just remains there for life as a "fossil" of the past infection and goes to the grave with the individual it's stuck in. All of us almost certainly contain countless such relics of the past viral infections we've fought off.
However... By chance this sometimes happens to a special cell in the body, a gametocyte cell that's one of the ones responsible for making sperm in males and egg cells in females, and if so subsequent sperm/eggs produced by that cell will contain copies of the "fossil" virus, since now it's just a portion of the entire DNA package of the cell. And once in a blue moon such a sperm or egg is lucky enough to be one of the few which participate in fertilization and are used to produce a child -- who will now inherit copies of the "fossilized" viral DNA in every cell of his/her body, since all are copied from the DNA of the original modified sperm/egg.
So now the "fossilized" viral DNA sequence will be passed on to *their* children, and their children's children, and so on. Through a process called neutral genetic drift, given enough time (it happens faster in smaller populations than large) the "fossil" viral DNA will either be flushed out of the population eventually, *or* by luck of the draw end up in every member of the population X generations down the road. It all depends on a roll of the genetic dice.
Due to the hurdles, "fossil" retroviral DNA strings (known by the technical name of "endogenous retroviruses") don't end up ubiquitous in a species very often, but it provably *does* happen. In fact, the Human DNA project has identified literally *thousands* of such fossilized "relics" of long-ago ancestral infections in the human DNA.
And several features of these DNA relics can be used to demonstrate common descent, including their *location*. The reason is that retroviruses aren't very picky about where their DNA gets inserted into the host DNA. Even in an infection in a *single* individual, each infected cell has the retroviral DNA inserted into different locations than any other cell. Because the host DNA is so enormous (billions of basepairs in humans, for example), the odds of any retroviral insertion event matching the insertion location of any other insertion event are astronomically low. The only plausible mechanism by which two individuals could have retroviral DNA inserted into exactly the same location in their respective DNAs is if they inherited copies of that DNA from the same source -- a common ancestor.
Thus, shared endogenous retroviruses between, say, ape and man is almost irrefutable evidence that they descended from a common ancestor. *Unless* you want to suggest that they were created separately, and then a virus they were both susceptible to infected both a man and an ape in EXACTLY the same location in their DNAs (the odds of such a match by luck are literally on the order of 1,000,000,000,000 to 1...), *and* that the infections both happened in their gametocyte cells (combined odds on the order of 1,000,000 to 1) *and* that the one particular affected gametocyte is the one which produces the egg or sperm which is destined to produce an offspring (*HUGE* odds against), and *then* the resulting modified genome of the offspring becomes "fixed" in each respective population (1 out of population_size^squared)...
Then repeat that for *each* shared endogenous retrovirus (there are many) you'd like to claim was acquired independently and *not* from a shared ancestor...
Finally, you'd have to explain why, for say species A, B, and C, the pattern of shared same-location retroviruses is always *nested*, never *overlapped*. For example, all three will share some retroviruses, then A and B will both share several more, but if so then B *never* shares one with C that A doesn't also have (or at least remnants of).
In your "shared infection due to genetic similarities" suggestion, even leaving aside the near statistical impossibility of the infections leaving genetic "scars" in *exactly* the same locations in independent infections, one would expect to find cases of three species X, Y, and Z, where the degree of similarity was such that Y was "between" X and Z on some similarity scale, causing the same disease to befall X and Y but not Z, and another disease to affect Y and Z but not X. And yet, we don't find this in genetic markers. The markers are found in nested sequence, which is precisely what we would expect to see in cases of inheritance from common ancestry.
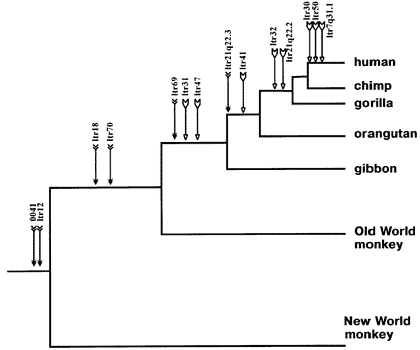
Here, for example, is an ancestry tree showing the pattern of shared same-location endogenous retroviruses of type HERV-K among primates:
This is just a partial list for illustration purposes -- there are many more.
Each labeled arrow on the chart shows an ERV shared in common by all the branches to the right, and *not* the branches that are "left-and-down". This is the pattern that common descent would make. And common descent is the *only* plausible explanation for it. Furthermore, similar findings tie together larger mammal groups into successively larger "superfamilies" of creatures all descended from a common ancestor.
Any presumption of independent acquisition is literally astronomically unlikely. And "God chose to put broken relics of viral infections that never actually happened into our DNA and line them up only in patterns that would provide incredibly strong evidence of common descent which hadn't actually happened" just strains credulity (not to mention would raise troubling questions about God's motives for such a misleading act).
Once again, the evidence for common descent -- as opposed to any other conceivable alternative explanation -- is clear and overwhelming.
Wait, want more? Endogenous retroviruses are just *one* type of genetic "tag" that makes perfect sense evolutionary and *no* sense under any other scenario. In addition to ERV's, there are also similar arguments for the patterns across species of Protein functional redundancies, DNA coding redundancies, shared Processed pseudogenes, shared Transposons (including *several* independent varieties, such as SINEs and LINEs), shared redundant pseudogenes, etc. etc. Here, for example, is a small map of shared SINE events among various mammal groups:
Like ERV's, any scenario which suggests that these shared DNA features were acquired separately strains the laws of probability beyond the breaking point, but they make perfect sense from an evolutionary common-descent scenario. In the above data, it is clear that the only logical conclusion is that, for example, the cetaceans, hippos, and ruminants shared a common ancestor, in which SINE events B and C entered its DNA and then was passed on to its descendants, yet this occurred after the point in time where an earlier common ancestor had given rise both to that species, and to the lineage which later became pigs.
And this pattern (giving the *same* results) is repeated over and over and over again when various kinds of molecular evidence from DNA is examined in detail.
The molecular evidence for evolution and common descent is overwhelming. The only alternative is for creationists to deny the obvious and say, "well maybe God decided to set up all DNA in *only* ways that were consistent with an evolutionary result even though He'd have a lot more options open to him, even including parts which by every measure are useless and exactly mimic copy errors, ancient infections, stutters, and other garbage inherited from nonexistent shared ancestors"...
And since a couple times when I've posted this, clueless anti-evolutionists have mistaken their presumptions for reality and tried to declare that ERV insertion or analysis was just "speculative", here are a few relevant papers to chew on establishing their validity:
Human-specific integrations of the HERV-K endogenous retrovirus family
Endogenous retroviruses in the human genome sequence
Constructing primate phylogenies from ancient retrovirus sequences
Human L1 Retrotransposition: cis Preference versus trans Complementation
Identification, Phylogeny, and Evolution of Retroviral Elements Based on Their Envelope Genes
HERVd: database of human endogenous retroviruses
Long-term reinfection of the human genome by endogenous retroviruses
Insertional polymorphisms of full-length endogenous retroviruses in humans
Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans
The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease
A rare event of insertion polymorphism of a HERV-K LTR in the human genome
Demystified . . . Human endogenous retroviruses
Retroviral Diversity and Distribution in Vertebrates
Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene
Genomic Organization of the Human Endogenous Retrovirus HERV-K(HML-2.HOM) (ERVK6) on Chromosome 7
Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H
And, as usual, that's just the tiniest *tip* of the iceberg. My PubMed searches on endogenous retroviruses turned up over a *thousand* papers. These are just some of the more useful ones.
Humans have 23 pairs of chromosomes ---chimps and gorillas have 24 pairs. How many pairs of chromosomes did the "common ancestor" have? Was it 23 or 24 pairs? How do you "evolve" missing or added chromosomes ---that would happen all at one time.And:The common ancestor had 24 chromosomes.
If you look at the gene sequences, you'll find that Chromosome 2 in humans is pretty much just 2 shorter chimpanzee chromosomes pasted end-to-end, with perhaps a slight bit of lost overlap:
(H=Human, C=Chimpanzee, G=Gorilla, O=Orangutan)
Somewhere along the line, after humans split off from the other great apes, or during the split itself, there was an accidental fusion of two chromosomes, end-to-end. Where there used to be 24 chromosomes, now there were 23, but containing the same total genes, so other than a "repackaging", the DNA "instructions" remained the same.
If a chimpanzee gives birth to a creature with 23 chromosomes, that offspring isn't going to be a well-formed chimpanzee able to survive well.
It is if the same genes are present, which they would be in the case of a chromosome fusion.
Evolve would imply the genetic material changes little by little --not some big loss of two chromosomes at once but I don't see how they'd go away gene by gene.
Tacking two chromosomes together end-to-end is not a "big loss" of genes, and it really is a "little by little" change in the total genetic code. It's just been "regrouped" a bit. Instead of coming in 24 "packages", it's now contained in 23, but the contents are the same.
So how, you might ask, would the chromosomes from the first 23-chromosome "fused" individual match up with the 24 chromosomes from its mate when it tried to produce offspring? Very well, thanks for asking. The "top half" of the new extra-long Chromosome 2 would adhere to the original chromosome (call it "2p") from which it was formed, and likewise for the "bottom half" which would adhere to the other original shorter chromosome (call it "2q"). In the picture above, imagine the two chimp chromosomes sliding over to "match up" against the human chromosome. The chimp chromosomes would end up butting ends with each other, or slightly overlapping in a "kink", but chromosomes have overcome worse mismatches (just consider the XY pair in every human male -- the X and the Y chromosome are *very* different in shape, length, and structure, but they still pair up).
In fact, the "rubbing ends" of the matched-up chimp chromosomes, adhering to the double-long human-type chromosome, would be more likely to become fused together themselves.
For studies in which recent chromosome fusions have been discovered and found not to cause infertility, see:
Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Hauffe HC, Searle JB Department of Zoology, University of Oxford, Oxford OX1 3PS, United Kingdom. hauffe@novanet.itIn that last reference, the Przewalski horse, which has 33 chromosomes, and the domestic horse, with 32 chromosomes (due to a fusion), are able to mate and produce fertile offspring.An observed chromosome fusion: Hereditas 1998;129(2):177-80 A new centric fusion translocation in cattle: rob (13;19). Molteni L, De Giovanni-Macchi A, Succi G, Cremonesi F, Stacchezzini S, Di Meo GP, Iannuzzi L Institute of Animal Husbandry, Faculty of Agricultural Science, Milan, Italy.
J Reprod Fertil 1979 Nov;57(2):363-75 Cytogenetics and reproduction of sheep with multiple centric fusions (Robertsonian translocations). Bruere AN, Ellis PM
J Reprod Fertil Suppl 1975 Oct;(23):356-70 Cytogenetic studies of three equine hybrids. Chandley AC, Short RV, Allen WR.
Meanwhile, the question may be asked, how do we know that the human Chromosome 2 is actually the result of a chromsome fusion at/since a common ancestor, and not simply a matter of "different design"?
Well, if two chromsomes accidentally merged, there should be molecular remnants of the original chromosomal structures (while a chromosome designed from scratch would have no need for such leftover "train-wreck" pieces).
Ends of chromosomes have characteristic DNA base-pair sequences called "telomeres". And there are indeed remnants of telomeres at the point of presumed fusion on human Chromosome 2 (i.e., where the two ancestral ape chromosomes merged end-to-end). If I may crib from a web page:
Telomeres in humans have been shown to consist of head to tail repeats of the bases 5'TTAGGG running toward the end of the chromosome. Furthermore, there is a characteristic pattern of the base pairs in what is called the pre-telomeric region, the region just before the telomere. When the vicinity of chromosome 2 where the fusion is expected to occur (based on comparison to chimp chromosomes 2p and 2q) is examined, we see first sequences that are characteristic of the pre-telomeric region, then a section of telomeric sequences, and then another section of pre-telomeric sequences. Furthermore, in the telomeric section, it is observed that there is a point where instead of being arranged head to tail, the telomeric repeats suddenly reverse direction - becoming (CCCTAA)3' instead of 5'(TTAGGG), and the second pre-telomeric section is also the reverse of the first telomeric section. This pattern is precisely as predicted by a telomere to telomere fusion of the chimpanzee (ancestor) 2p and 2q chromosomes, and in precisely the expected location. Note that the CCCTAA sequence is the reversed complement of TTAGGG (C pairs with G, and T pairs with A).Another piece of evidence is that if human Chromosome 2 had formed by chromosome fusion in an ancestor instead of being designed "as is", it should have evidence of 2 centromeres (the "pinched waist" in the picture above -- chromosomes have centromeres to aid in cell division). A "designed" chromosome would need only 1 centromere. An accidentally "merged" chromosome would show evidence of the 2 centromeres from the two chromosomes it merged from (one from each). And indeed, as documented in (Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M, Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet 1992 May;89(2):247-9), the functional centromere found on human Chromosome 2 lines up with the centromere of the chimp 2p chromosome, while there are non-functional remnants of the chimp 2q centromere at the expected location on the human chromosome.As an aside, the next time some creationist claims that there is "no evidence" for common ancestry or evolution, keep in mind that the sort of detailed "detective story" discussed above is repeated literally COUNTLESS times in the ordinary pursuit of scientific research and examination of biological and other types of evidence. Common ancestry and evolution is confirmed in bit and little ways over and over and over again. It's not just something that a couple of whacky anti-religionists dream up out of thin air and promulgate for no reason, as the creationists would have you believe.
[The poster known as Mr. LLLICHY wrote:] Here is that Vitamin C dataAnd:After discovering this same data on another thread along with more discussion than has appeared here (I've taken the liberty of pinging the participants of that discussion), I see what the "mystery" is supposed to be -- it's supposed be why did some sites have multiple mutations while (small) stretches of other sites had none? In other words, why do the mutations appear clustered?
(You know, it would really help if people explained their points and questions in more detail, instead of leaving people to guess what the poster was thinking...)
[LLLICHY wrote:] "U238" that decays thrice, pretty good trick when there is "U238" that does not decay at all in 50,000,000 years.
Actually, no site had mutations "thrice". Three different bases at a given site is only *two* mutations (one original base, plus two mutations from it to something else).
Here's the "mutation map" from the actual DNA data:
--1-12--1-1-1-1--------1112112--1---1-11-1--------1 ALL/nNo mutations ("-") in about half the sites, one mutation at several (17) sites, two mutations at three sites.The first thing to keep in mind that random processes tend to "cluster" more than people expect anyway. People expect "randomness" to "spread out" somewhat evenly, but instead it's usually more "clumped", for statistical reasons that would be a diversion to go into right now. So "that looks uneven" isn't always a good indication that something truly is non-random.
If you don't believe me on that, I wrote a program which made 23 mutations totally at random on a 51-site sequence, then repeated the process to see what different random outcomes would look like:
10 X$=STRING$(51,"-") 20 FOR I=1 TO 23 30 J%=INT(RND*51)+1 40 C$=MID$(X$,J%,1) 50 IF C$="-" THEN MID$(X$,J%,1)="1" ELSE MID$(X$,J%,1)=CHR$(ASC(C$)+1) 60 NEXT I 70 PRINT X$ 80 GOTO 10Yeah, it's BASIC, so sue me. Here's a typical screenful of the results:-21---1---2---111----2-----2-1121-------1---1--11-1 -1--1--21-11---1-1--1-1---1----1---21-11111---11--- 3-11---3-----1-----11-2-1---1--1----3--2---1--1---- ---1-1--22--1-1--2-2111--1-1111---1------1-------1- ---32----1-11-1-----1---2-231----1------1-----11--1 ----2---21--1---4----1-------------11-1--111-11-211 11--1-1---1-----1--1------1----3111--1----111-2-1-2 1112---1-3-1----1-1-----1-1------121--111-------1-1 -111121--1----1----1-1-1-1-11-2---1-1-------1-111-- -----------11-1---11-11--------21----12211--1---131 --1-211-1-1----21--11-1-2----1--1----11---11-----11 12---1-13------------2---21-21---11-1-1-1--2------- -----2-1---1-1----21--11-11-1---111-1--111-----2--1 -----1-----1-1-1-1---1-2----11-21-11--1-111---1-21- ---11--1-1-122-1-1-1--1-----2-1-1-1-------1-1---111 --2--11----2--1---12-2----1-1---1-1--1--12----1-1-1 -111-1-----1-1----------1-21111--1-2-11-11-1----11- 11-1--211-1221-----1--1-----11--1-2-1----------11-- -----1-12-11---2-1---11--1-2--1----11---111-1----11 11----1--12---12----1---31---1-11----2--1-11-1----- ---1--111-1--1-1-111----1-21----1-1-3---1------2--1 -2-11----1-1------1------2-1-1--111-111-1-1----1111 1--1--1-1---1-111111--2--1-1------112----2---11----Notice how oddly "clustered" most of them look, including one run which left a 13-site stretch "absolutely untouched", contrary to intuition (while having *4* mutations at a single site!)Frankly, I don't see anything in the real-life DNA mutation map which looks any different from these truly random runs. Random events tend to cluster more than people expect. That solves the "mystery" right there.
Also, there may be a selection factor -- the GLO gene is a *lot* bigger than this. One has to wonder if this small 51-bp section was presented just because it was the one that looked "least random". That would be a no-no, since one can always hand-select the most deviant subset out of larger sample in order to artificially skew the picture.
However, since there are some interesting evolutionary observations to be made, let's look at that DNA data again, slightly rearranged:
TAC CCC GTG GAG GTG CGC TTC ACT CGG GCG GAC GAC ATC CTG CTG AGC CCC PIG TAC CCC GTG GAG GTA CGC TTC ACT CGC GGG GAC GAC ATC CTG CTG AGC CCC BOS TAC CCC GTA GAG GTG CGC TTC ACC CGA GGC GAT GAC ATT CTG CTG AGC CCC RAT TAC CCC GTG GAG GTG CGC TTC ACC CGA GGT GAT GAC ATC CTG CTG AGC CCG MOUSE TAC CCT GTG GGG GTG CGC TTC ACC CGG GGG GAC GAC ATC CTG CTG AGC CCC GUIN PIG TAC CTG GTG GGG GTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC HUMAN TAC CTG GTG GGG CTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC CHIMPANZEE TAC CCG GTG GGG GTG CGC TTC ACC CAG AG* GAT GAC GTC CTA CTG AGC CCC ORANGUTAN TAA CCG GTG GGG GTG CGC TTC ACC CAA GG* GAT GAC ATC ATA CTG AGC CCC MACAQUEHere I've put spaces between codons, and clustered the closely-related species together: pig/cow as ungulates, rat/mouse for their obvious relationship, guinea pig right below them but separated because of the pseudogene nature of its GLO gene, then primates all in a group, with man's closest relative, the chimp, immediately below him, followed by the more distant orangutan, and the even more distant macaque. Also note that the top four have "working" GLO genes, and the bottom five have "broken" GLO pseudogenes.First, let's consider just the four species with working GLO genes. Evolution predicts that even over large periods of time, these genes will be "highly conserved", with natural selection weeding out mutations that could "break" the gene. Note that the mutations will still have occurred in individuals of the population, but natural selection will "discourage" that mutation from spreading into the general population.
And before we go any further, let's talk about the "universal genetic code". In all mammals (indeed, in almost all living organisms), each triplet of DNA sites cause a particular amino acid to be formed. The mapping of triplets (called "codons") to amino acids is as follows:
Second Position of Codon T C A G F
i
r
s
t
P
o
s
i
t
i
o
nT
TTT Phe [F] TTC Phe [F] TTA Leu [L] TTG Leu [L]
TCT Ser [S] TCC Ser [S] TCA Ser [S] TCG Ser [S]
TAT Tyr [Y] TAC Tyr [Y] TAA Ter [end] TAG Ter [end]
TGT Cys [C] TGC Cys [C] TGA Ter [end] TGG Trp [W]
T C A G T
h
i
r
d
P
o
s
i
t
i
o
nC
CTT Leu [L] CTC Leu [L] CTA Leu [L] CTG Leu [L]
CCT Pro [P] CCC Pro [P] CCA Pro [P] CCG Pro [P]
CAT His [H] CAC His [H] CAA Gln [Q] CAG Gln [Q]
CGT Arg [R] CGC Arg [R] CGA Arg [R] CGG Arg [R]
T C A G A
ATT Ile [I] ATC Ile [I] ATA Ile [I] ATG Met [M]
ACT Thr [T] ACC Thr [T] ACA Thr [T] ACG Thr [T]
AAT Asn [N] AAC Asn [N] AAA Lys [K] AAG Lys [K]
AGT Ser [S] AGC Ser [S] AGA Arg [R] AGG Arg [R]
T C A G G
GTT Val [V] GTC Val [V] GTA Val [V] GTG Val [V]
GCT Ala [A] GCC Ala [A] GCA Ala [A] GCG Ala [A]
GAT Asp [D] GAC Asp [D] GAA Glu [E] GAG Glu [E]
GGT Gly [G] GGC Gly [G] GGA Gly [G] GGG Gly [G]
T C A G (The above table imported from http://psyche.uthct.edu/shaun/SBlack/geneticd.html, which also has a nice introduction to the genetic code.)
Another version of the same table with nifty Java features and DNA database lookups can be found here.
The thing which is most relevant to the following discussion is the fact that most of the genetic codes are "redundant" -- more than one codon (triplet) encodes to exactly the same amino acid. This means that even in genes which are required for the organism, certain basepair mutations make absolutely no difference if the change is from one codon which maps into amino acid X to another codon which still maps into amino acid X. (This fact allows certain kinds of evolutionary "tracers" to be "read" from the DNA, as described here).
Now back to our DNA data. The redundancy in the genetic code means that some basepair sites will have more "degrees of freedom" than others (i.e., ways in which they can mutate without disrupting the gene's biological function in any way). Let's look at the four species with working GLO genes again:
TAC CCC GTG GAG GTG CGC TTC ACT CGG GCG GAC GAC ATC CTG CTG AGC CCC PIG TAC CCC GTG GAG GTA CGC TTC ACT CGC GGG GAC GAC ATC CTG CTG AGC CCC BOS TAC CCC GTA GAG GTG CGC TTC ACC CGA GGC GAT GAC ATT CTG CTG AGC CCC RAT TAC CCC GTG GAG GTG CGC TTC ACC CGA GGT GAT GAC ATC CTG CTG AGC CCG MOUSE T T T A T A T T T A T C C T T T T T T T T A A A A A C A A A A A G C G G G G G C C C --- --- --1 --- --1 --- --- --1 --2 -12 --1 --- --1 --- --- --- --1Under each site of the mouse DNA, I've listed the "alternative" bases which could be be substituted for the mouse base at that site WITHOUT ALTERING THE GENE'S FUNCTION (because of genetic code redundancy). And under that I show the "mutation map" of just those four species.Note that most of the "alternative" bases are in the third base of each codon, *and* that this is where all but one of the mutations have appeared. This is because these were the sites which were "free" to mutate in the way they did, because the mutation was genetically neutral. That doesn't mean that the first and second sites of each codon were immune from mutation, it's just that when mutations did occur at those sites, natural selection weeded them out quickly because they most likely "broke" the GLO gene for the individuals which received that mutuation. What we see above is the results after natural selection has already "filtered" the undesirable mutations and left the ones which "do no harm".
Additionally, the two sites which have mutated twice (i.e. have a "2" in the mutation map) are ones which had more "allowable" mutations. Also note that the sites which had the fewest allowable alternatives (only one alternate letter allowed) didn't have any mutations fix at those sites, which is unsurprising since a "safe" mutation would be less likely to occur there versus a site that "allowed" two or three alternatives.
All this is as predicted by evolutionary theory, you'll note.
It also explains the one anomoly of the original mutation map, which is that the mutation counts do tend to be higher at the third base of a codon.
However... What about the one exception? The pig DNA has had one mutation at a site which does not encode to exactly the same amino acid (which is the case for *all* the other ones). In the pig DNA, the GGG codon (mapping to Glycine) has changed to a GCG codon (mapping to Alanine). What's up with that? Well, one of two things. First and most likely, just as base values in codons have a built-in redundancy, so do the amino acids which make up the proteins which result from the DNA templates. In other words, certain amino acids can be substituted for other ones at some sites in given proteins without making any functional difference. (This "protein functional redundancy" also has implications for "evolutionary tracer" analysis, see here.) That may well be the case for Alanine versus Glycine in the GLO protein, but I'm not enough of a biochemist to be able to say. The other option is that it *does* make some difference in the function of the pig GLO protein, but not enough to "break" the vitamin-C synthesis (as proven by the fact that pigs *can* synthesize vitamin C). So one way or another, it's not a deal-breaker even though pig GLO will not be 100% identical to cow/mouse/rat GLO. It's yet another "allowable" mutation.
More interesting evolutionary observations: The number of mutational differences between pig/cow is 3, the number between mouse/rat is 4, and the difference between rat/cow is 7 -- all roughly as one would expect from the evolutionary relatedness of these animals (cows/pigs and rats/mice are each closer to each other than the rodents are to the ungulates).
Now let's take a close look at the guinea pig:
TAC CCT GTG GGG GTG CGC TTC ACC CGG GGG GAC GAC ATC CTG CTG AGC CCC GUIN PIG --- --1 --- -1- --- --- --- --- --1 --1 --1 --- --- --- --- --- ---The "mutation map" under the guinea pig DNA is compared to the mouse DNA. Fascinating: Note that four of the five mutations are in the third base of a codon, *and* are of the type "allowed" by the genetic code redundancy. This indicates strongly that most of the evolutionary divergence between guinea pigs and mice likely occurred while the guinea pig's ancestors still had a working GLO gene. This is the sort of prediction implied by the evolutionary theory which could be cross-checked by further research of various types, and if verified, would be yet further confirmation that evolutionary theory is likely correct. So far, evolutionary theory has been subjected to literally countless tests like this, large and small, and the vast majority of results have confirmed the evolutionary prediction. This track record is hard to explain if evolution is an invalid theory, as some assert...Finally, let's look over the primate DNA and mutation map (relative to each other):
TAC CTG GTG GGG GTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC HUMAN TAC CTG GTG GGG CTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC CHIMPANZEE TAC CCG GTG GGG GTG CGC TTC ACC CAG AG* GAT GAC GTC CTA CTG AGC CCC ORANGUTAN TAA CCG GTG GGG GTG CGC TTC ACC CAA GG* GAT GAC ATC ATA CTG AGC CCC MACAQUE --1 -1- --- --- 1-1 --- --- --- 111 1-- --- --- 1-- 1-- --- --- ---Evolutionary theory predicts that because the GLO gene is "broken" in primates (i.e. is a pseudogene), mutations in it are highly likely to be neutral (i.e., make no difference, since it can't get much more broken), and thus mutations are just as likely to accumulate at any site as any other. Is that what we see? Yup. There's no obvious pattern to the mutations between primates in the above mutation map, and unlike the pig/cow/mouse/rat mutation map, the mutations aren't predominantly at the "safer" third base of a codon, nor of a type that would be "safe". In fact, one base has vanished entirely, but no biggie, the gene's already broken.Also, although primates share a more recent common ancestor than cows/pigs/mice/rats, note that they've already racked up almost as many relative mutations as the cow/pig/mouse/rat DNA. This too is just as evolutionary theory predicts, because many mutations in a functional gene (GLO in this case) will be "non-safe" and weeded out by natural selection, making for a slower mutation fixation rate overall than in a pseudogene (as GLO is in primates) where natural selection doesn't "care" about the vast majority of mutations since *most* are neutral. So pseudogenes accumulate mutations faster than functional genes (even though rate of mutation *occurence* in both are likely the same).
Finally, note that there are ZERO mutational differences between the human DNA and the chimpanzee DNA, our nearest living relative.
I also see some interesting implications in the DNA sequences concerning which specific mutation fixed during what branch of the common-descent evolutionary tree for all the species represented, but reconstructing that would not only take another couple hours, at least, but would be a major bear to code in HTML, since I'd have to draw trees with annotations on the nodes... Bleugh.
In any case, I hope I've clarified some of the methods by which biologists find countless confirmations of evolution in DNA data. This is just a "baby" example, and to be more statistically valid would have to be done over much vaster sections of DNA sequences, but my intent was to demonstrate some of the concepts.
And if such a small amount of DNA as this can make small confirmations of evolutionary predictions, imagine the amount of confirmation from billion-basepair DNA data from each species compared across thousands of species... The amount of confirmatory discoveries for evolution from DNA analysis has already been vast, and promises to only grow in the future. For an overview of some of the different lines of evidence being studied, see The Journal of Molecular Evolution -- abstracts of all articles, current and back issues, can be browsed free online.
And:Fossil Hominids: The Evidence for Human Evolution.
29 Evidences for Macroevolution -- Part 1: The Unique Universal Phylogenetic Tree
Analysis of the human Alu Ye lineage
Molecular and temporal characteristics of human retropseudogenes.
Serine hydroxymethyltransferase pseudogene, SHMT-ps1: a unique genetic marker of the order primates
Insertions and duplications of mtDNA in the nuclear genomes of Old World monkeys and hominoids
Fixation times of retroposons in the ribosomal DNA spacer of human and other primates
The emergence of new DNA repeats and the divergence of primates
Genetic diversity at class II DRB loci of the primate MHC
Structure and evolution of human and African ape rDNA pseudogenes
Accelerated Evolution of the ASPM Gene Controlling Brain Size Begins Prior to Human Brain ExpansionEtc., Etc., Etc. How many more thousands of papers would you like to see covering the common ancestry that the author claims there is "no evidence supporting"?Abstract: Primary microcephaly (MCPH) is a neurodevelopmental disorder characterized by global reduction in cerebral cortical volume. The microcephalic brain has a volume comparable to that of early hominids, raising the possibility that some MCPH genes may have been evolutionary targets in the expansion of the cerebral cortex in mammals and especially primates. Mutations in ASPM, which encodes the human homologue of a fly protein essential for spindle function, are the most common known cause of MCPH. Here we have isolated large genomic clones containing the complete ASPM gene, including promoter regions and introns, from chimpanzee, gorilla, orangutan, and rhesus macaque by transformation-associated recombination cloning in yeast. We have sequenced these clones and show that whereas much of the sequence of ASPM is substantially conserved among primates, specific segments are subject to high Ka/Ks ratios (nonsynonymous/synonymous DNA changes) consistent with strong positive selection for evolutionary change. The ASPM gene sequence shows accelerated evolution in the African hominoid clade, and this precedes hominid brain expansion by several million years. Gorilla and human lineages show particularly accelerated evolution in the IQ domain of ASPM. Moreover, ASPM regions under positive selection in primates are also the most highly diverged regions between primates and nonprimate mammals. We report the first direct application of TAR cloning technology to the study of human evolution. Our data suggest that evolutionary selection of specific segments of the ASPM sequence strongly relates to differences in cerebral cortical size.Identification of paralogous HERV-K LTRs on human chromosomes 3, 4, 7 and 11 in regions containing clusters of olfactory receptor genesAbstract: A locus harboring a human endogenous retroviral LTR (long terminal repeat) was mapped on the short arm of human chromosome 7 (7p22), and its evolutionary history was investigated. Sequences of two human genome fragments that were homologous to the LTR-flanking sequences were found in human genome databases: (1) an LTR-containing DNA fragment from region 3p13 of the human genome, which includes clusters of olfactory receptor genes and pseudogenes; and (2) a fragment of region 21q22.1 lacking LTR sequences. PCR analysis demonstrated that LTRs with highly homologous flanking sequences could be found in the genomes of human, chimp, gorilla, and orangutan, but were absent from the genomes of gibbon and New World monkeys. A PCR assay with a primer set corresponding to the sequence from human Chr 3 allowed us to detect LTR-containing paralogous sequences on human chromosomes 3, 4, 7, and 11. The divergence times for the LTR-flanking sequences on chromosomes 3 and 7, and the paralogous sequence on chromosome 21, were evaluated and used to reconstruct the order of duplication events and retroviral insertions. (1) An initial duplication event that occurred 14-17 Mya and before LTR insertion - produced two loci, one corresponding to that located on Chr 21, while the second was the ancestor of the loci on chromosomes 3 and 7. (2) Insertion of the LTR (most probably as a provirus) into this ancestral locus took place 13 Mya. (3) Duplication of the LTR-containing ancestral locus occurred 11 Mya, forming the paralogous modern loci on Chr 3 and 7.Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter fluxAbstract: The enzyme glutamate dehydrogenase (GDH) is important for recycling the chief excitatory neurotransmitter, glutamate, during neurotransmission. Human GDH exists in housekeeping and brain-specific isotypes encoded by the genes GLUD1 and GLUD2, respectively. Here we show that GLUD2 originated by retroposition from GLUD1 in the hominoid ancestor less than 23 million years ago. The amino acid changes responsible for the unique brain-specific properties of the enzyme derived from GLUD2 occurred during a period of positive selection after the duplication event.A uniquely human consequence of domain-specific functional adaptation in a sialic acid–binding receptorAbstract: Most mammalian cell surfaces display two major sialic acids (Sias), N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc). Humans lack Neu5Gc due to a mutation in CMP-Neu5Ac hydroxylase, which occurred after evolutionary divergence from great apes. We describe an apparent consequence of human Neu5Gc loss: domain-specific functional adaptation of Siglec-9, a member of the family of sialic acid–binding receptors of innate immune cells designated the CD33-related Siglecs (CD33rSiglecs). Binding studies on recombinant human Siglec-9 show recognition of both Neu5Ac and Neu5Gc. In striking contrast, chimpanzee and gorilla Siglec-9 strongly prefer binding Neu5Gc. Simultaneous probing of multiple endogenous CD33rSiglecs on circulating blood cells of human, chimp, or gorilla suggests that the binding differences observed for Siglec-9 are representative of multiple CD33rSiglecs. We conclude that Neu5Ac-binding ability of at least some human CD33rSiglecs is a derived state selected for following loss of Neu5Gc in the hominid lineage. These data also indicate that endogenous Sias (rather than surface Sias of bacterial pathogens) are the functional ligands of CD33rSiglecs and suggest that the endogenous Sia landscape is the major factor directing evolution of CD33rSiglec binding specificity. Exon-1-encoded Sia-recognizing domains of human and ape Siglec-9 share only 93–95% amino acid identity. In contrast, the immediately adjacent intron and exon 2 have the 98–100% identity typically observed among these species. Together, our findings suggest ongoing adaptive evolution specific to the Sia-binding domain, possibly of an episodic nature. Such domain-specific divergences should also be considered in upcoming comparisons of human and chimpanzee genomes.Lineage-Specific Gene Duplication and Loss in Human and Great Ape EvolutionAbstract: Given that gene duplication is a major driving force of evolutionary change and the key mechanism underlying the emergence of new genes and biological processes, this study sought to use a novel genome-wide approach to identify genes that have undergone lineage-specific duplications or contractions among several hominoid lineages. Interspecies cDNA array-based comparative genomic hybridization was used to individually compare copy number variation for 39,711 cDNAs, representing 29,619 human genes, across five hominoid species, including human. We identified 1,005 genes, either as isolated genes or in clusters positionally biased toward rearrangement-prone genomic regions, that produced relative hybridization signals unique to one or more of the hominoid lineages. Measured as a function of the evolutionary age of each lineage, genes showing copy number expansions were most pronounced in human (134) and include a number of genes thought to be involved in the structure and function of the brain. This work represents, to our knowledge, the first genome-wide gene-based survey of gene duplication across hominoid species. The genes identified here likely represent a significant majority of the major gene copy number changes that have occurred over the past 15 million years of human and great ape evolution and are likely to underlie some of the key phenotypic characteristics that distinguish these species.Sequence Variation Within the Fragile X LocusAbstract: The human genome provides a reference sequence, which is a template for resequencing studies that aim to discover and interpret the record of common ancestry that exists in extant genomes. To understand the nature and pattern of variation and linkage disequilibrium comprising this history, we present a study of ~31 kb spanning an ~70 kb region of FMR1, sequenced in a sample of 20 humans (worldwide sample) and four great apes (chimp, bonobo, and gorilla). Twenty-five polymorphic sites and two insertion/deletions, distributed in 11 unique haplotypes, were identified among humans. Africans are the only geographic group that do not share any haplotypes with other groups. Parsimony analysis reveals two main clades and suggests that the four major human geographic groups are distributed throughout the phylogenetic tree and within each major clade. An African sample appears to be most closely related to the common ancestor shared with the three other geographic groups. Nucleotide diversity, [pi], for this sample is 2.63 ± 6.28 × 10-4. The mutation rate, [mu], is 6.48 × 10-10 per base pair per year, giving an ancestral population size of ~6200 and a time to the most recent common ancestor of ~320,000 ± 72,000 per base pair per year. Linkage disequilibrium (LD) at the FMR1 locus, evaluated by conventional LD analysis and by the length of segment shared between any two chromosomes, is extensive across the region.Structural and evolutionary analysis of the two chimpanzee alpha-globin mRNAsAbstract: Two distinct alpha-globin mRNAs were detected in chimpanzee reticulocyte mRNA using a primer extension assay. DNA copies of these two mRNAs were cloned in the bacterial plasmid pBR322, and their sequence was determined. The two alpha-globin mRNAs have obvious structural homology to the two human alpha-globin mRNAs, alpha 1 and alpha 2. Comparison of the two chimpanzee alpha-globin mRNAs to each other and to their corresponding human counterparts revealed evidence of a recent gene conversion in the human alpha-globin complex and a marked heterogeneity in the rate of structural divergence within the alpha-globin gene.Differential Alu Mobilization and Polymorphism Among the Human and Chimpanzee LineagesAbstract: Alu elements are primate-specific members of the SINE (short interspersed element) retroposon family, which comprise10% of the human genome. Here we report the first chromosomal-level comparison examining the Alu retroposition dynamics following the divergence of humans and chimpanzees. We find a twofold increase in Alu insertions in humans in comparison to the common chimpanzee (Pan troglodytes). The genomic diversity (polymorphism for presence or absence of the Alu insertion) associated with these inserts indicates that, analogous to recent nucleotide diversity studies, the level of chimpanzee Alu diversity is
1.7 times higher than that of humans. Evolutionarily recent Alu subfamily structure differs markedly between the human and chimpanzee lineages, with the major human subfamilies remaining largely inactive in the chimpanzee lineage. We propose a population-based model to account for the observed fluctuation in Alu retroposition rates across primate taxa.
In fact, the answer is a categorical No.
The author is welcome to believe any blatant falsehood he wants, including this one.
Australopithecines, for example, were simply extinct apes that in a few anatomical areas differed from living apes.
If some of them walked bipedally to a greater degree than living apes, this does not constitute evidence that apes evolved into humans
That by itself wouldn't, of course, but the author sort of "forgets" to mention that there is *far* more to the analysis than just walking bipedally. OOPS!
- it just means that some ancient apes were different from living apes.
...only if one totally ignores such a vast amount of *additional* evidence, along multiple independently cross-confirming lines, as to make one's case by grossly misrepresenting reality...
Read all of the above post, and follow the links and read *them*, read some of the linked primary literature, then read Patrick Henry's "list o' links", then read all of the pages on these two indexes: Talk-Origins FAQ and Talk-Origins Must-Read Files. Finally, go over this List of Creationist Claims" (all 200+ of them) -- you'll find that although many of them may sound plasible at first glance, upon closer examination the vast manority evaporate into fallacies, misrepresentations, misunderstandings, and often just outright lies.
I know that may seem like asking a lot, but that's a bare *minimum* of the background one should have before being rash enough to attempt to critique a very well-established field of science, much less try to characterize the amount and strength of its evidence, what it has or has not already researched and validated, what it has or has not already addressed and settled, and so on. All too many people think that they're well-equipped to attack a 150-year-old branch of science and all of its body of accumulated knowledge, evidence, and research, without actually spending time really *learning* the field first, basing their views about it on having read little more than some anti-evolution screeds, and picking up their "knowledge" of evolutionary biology from the cartoonish misrepresentation of the real thing as described in the anti-evolution material.
But the problem is that trying to "learn" about evolutionary biology from anti-evolution creationist sources is like trying to "learn" about conservatism from Michael Moore and Cindy Sheehan -- and for exactly the same reasons.