Posted on 06/30/2013 9:56:45 PM PDT by neverdem

Researchers at the Lawrence Berkeley National Laboratory have imaged the atom-by-atom structure of the ribosome attached to a molecule that controls its motion for the first time, providing a step forward for the development of better antibiotics.
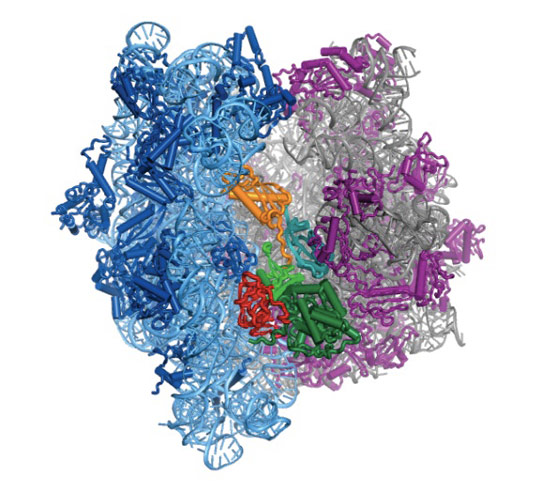
The above image may look like a tangle of squiggly lines, but you’re actually looking at a molecular machine called a ribosome. Its job is to translate DNA sequences into proteins, the workhorse compounds that sustain you and all living things.
The image is also a milestone. It’s the first time the atom-by-atom structure of the ribosome has been seen as it’s attached to a molecule that controls its motion. That’s big news if you’re a structural biologist.
But there’s another way to look at this image, one that anyone who’s suffered a bacterial infection can appreciate. The image is also a roadmap to better antibiotics. That’s because this particular ribosome is from a bacterium. And somewhere in its twists and turns could be a weakness that a new antibiotic can target.
“We’re in an arms race with the resistance mechanisms of bacteria,” says Jamie Cate, a staff scientist in Berkeley Lab’s Physical Biosciences Division and a professor of biochemistry, biophysics and structural biology at UC Berkeley.
“The better we understand how bacterial ribosomes work, the better we can come up with new ways to interfere with them,” he adds.
Cate developed the structure with UC Berkeley’s Arto Pulk. Their work is described in the June 28 issue of the journal Science.
Their image is the latest advance in the push for more effective antibiotics. The goal is new drugs that kill the bacteria that make us sick, stay one step ahead of their resistance mechanisms, and leave our beneficial bacteria alone.
One way to do this is to get to know the bacterial ribosome inside and out. Many of today’s antibiotics target ribosomes. A better understanding of how ribosomes function will shed light on how these antibiotics work. This could also lead to even “smarter” molecules that quickly target and disable a pathogen’s ribosomes without affecting friendly bacteria.
Cate and Pulk used protein crystallography beamlines at Berkeley Lab’s Advanced Light Source to create diffraction patterns that show how the ribosome’s molecules fit together. They then used computational modeling to combine these patterns into incredibly high-resolution images that describe the locations of the individual atoms.
The result is the colorful structure at the top of this article. Those blue and purple halves are ribosomes. They’re from E. coli bacteria, but they work in similar ways throughout nature. Ribosomes move along messenger RNA and interpret its genetic code into directions on how to stitch amino acids into proteins.
But sometimes ribosomes want to move backward, which isn’t good when you’re in the protein-making business. That’s where that yellow-red-green squiggle wedged between the two ribosome halves comes in. It’s elongation factor G. It acts like a ratchet and prevents the ribosome from slipping backward. It also pushes the ribosome forward when it’s sluggish.
Scientists knew that elongation factor G performs these jobs, but they didn’t know how. Now, with an atomic-scale structure in hand, they can study the chemical and molecular forces involved in this ratcheting process. Cate and Pulk found that the ratchet controls the ribosome’s motion by stiffening and relaxing over and over. This is the kind of insight that could lead to new ways to monkey-wrench the ribosome.
“To create better antibiotics, we need to learn how bacterial ribosomes work at the smallest scales, and this is a big step in that direction,” says Cate.
The National Institutes of Health and the National Cancer Institute supported the research. The U.S. Department of Energy provides support for the Advanced Light Source, where this research was conducted.
Publication: Arto Pulk, et al., “Control of Ribosomal Subunit Rotation by Elongation Factor G,” Science 28 June 2013: Vol. 340 no. 6140; DOI: 10.1126/science.1235970
Source: Dan Krotz, Lawrence Berkeley National Laboratory
Image: Lawrence Berkeley National Laboratory
And to think... It all “evolved” by vast amounts of randomness.
/s
Cancer Scientists Prove Long-Standing Theory on How Cancer Spreads hat tip yefragetuwrabrumuy
FReepmail me if you want on or off my combined microbiology/immunology ping list.
bookmark.
I remember a day long, long ago when I didn't mind the Feds funding such basic research.
Because it was basic RESEARCH, not some ego driven grant sucking fake research designed to prove how smart the lead researcher (and their grads students) are, and to fund the next two year cycle.
This is extremely impressive. Sounds like fun work.
Or it could lead to better bacteria.
Keep in mind that this marvelous molecular computational machine (along with a few billion other such machines in the cell, along with their chromosomal programming) accidentally “evolved” from a mud puddle. You know, pretty much like how the Intel i5 processor, Windows 7 and Linux accidentally evolved from some sand on the beach and some seawater.
Windows did, in fact, evolve from slime. Nobody in his right mind would create a “registry”.
Disclaimer: Opinions posted on Free Republic are those of the individual posters and do not necessarily represent the opinion of Free Republic or its management. All materials posted herein are protected by copyright law and the exemption for fair use of copyrighted works.