One of the greatest challenges in evolutionary biology is trying to piece together the history of life based on fragmentary evidence and limited knowledge of the forces at play — the rich tapestry of climate, geology and living things that forms the backdrop for evolution. How, for example, did the light-sensitive molecules that enable color vision come into being? Why did fluorescent corals start out green and then diversify into a rainbow of colors?
In the last decade, scientists in the emerging field of paleogenomics have developed new methods to begin answering some of these questions. By comparing the DNA sequences of many living organisms and working backward, they can infer ancient genomic history, such as when a gene evolved a new function. “A shared gene represents a fragment of information that has survived from the past,” said Betul Kacar, a research fellow at Harvard University. Researchers have been able to reconstruct a number of ancient proteins, such as those that detect steroid hormones, by tracing the path these molecules took to acquire their modern identities. Several years ago, Eric Gaucher, a biologist at the Georgia Institute of Technology in Atlanta, even resurrected a 700-million-year-old protein from E. coli.
Now, in a new twist on paleogenomics, Kacar has engineered that ancient protein into modern E. coli and tracked how the microbe adapted to it. The new approach, which Kacar presented yesterday at NASA’s Astrobiology Science Conference in Chicago, provides a more integrated view of the mechanisms of evolution — for example, how a protein’s position in a broader network influences its rate of change or how protein networks evolve as a whole. These kinds of insights will likely have practical implications as well, helping to predict how organisms might respond to climate change and other disturbances and enabling microbes to be more precisely engineered for medical and industrial uses.
Ancient History
Scientists have long been able to insert proteins from one organism into another, a basic component of genetic engineering. That’s how researchers engineer microbes to make fuels or medicines. But the results of these experiments can be unpredictable. “Sometimes it doesn’t work at all, and we don’t really understand why,” Gaucher said.

Courtesy of Eric Gaucher
Eric Gaucher, a biologist at the Georgia Institute of Technology, has reconstructed ancient versions of a number of different proteins.
A protein called elongation factor Tu (EF-Tu) is one such troublesome example. Despite being highly conserved (it’s present in all life on Earth) and essential (without it, cells die), it lacks flexibility. Putting a yeast or fruit-fly version of the EF-Tu protein into E. coli doesn’t work. “It’s one of the most highly conserved sequences in life, but it’s not swappable among life forms,” Gaucher said. “That’s odd.”
Kacar and Gaucher reasoned that perhaps an ancient version of the protein, derived from a 700-million-year-old ancestor of E. coli, would work better than a modern version from a different species. Gaucher compares it to dropping an English-speaking American into present-day Beijing or 1980s-era New York. The traveler would probably have an easier time navigating the American city than the Chinese one, where both language and culture are drastically different.
To deduce the structure of the ancestral version, Gaucher had compared the sequences of all known versions of the gene that produces the EF-Tu protein and used sophisticated computational and statistical methods to infer the gene’s most probable sequence. Kacar then synthesized that gene and inserted it into E. coli in place of the existing version. (She dubbed the hybrid the Rip strain, after Rip Van Winkle, because the gene has awoken from a 700-million-year sleep.)
The hybrid E. coli clearly suffered from the archaic component. The hybrids grew much more slowly than their normal counterparts, producing 25 percent fewer offspring. Just as a modern laptop would run poorly if its processor were replaced with an 1980s-era computer chip, the microbes’ modern molecular machinery simply wasn’t well suited to the ancient version of the protein. That’s because EF-Tu is a hub protein, interacting with 50 or more other proteins as it carries out its function in the cell. “When we put in the ancient form, we are undoubtedly disrupting some of those interactions,” Gaucher said. The partner proteins have undergone their own evolutionary changes over the last 700 million years — they’re designed to work with the laptop’s modern components, not the outdated chip.
Unlike a computer, however, the bacteria quickly bounced back. Kacar grew the hybrids in the lab, checking their growth rates and other factors every few hundred generations. Within a couple of months — about 500 generations — the hybrid E. coli were growing as well as their modern counterparts. These survivor strains must have evolved ways to overcome the problems caused by the outdated protein. But how?
Making Amends
Other experiments have inserted engineered proteins into novel environments. In trial after trial, the protein evolves via the exact same path, as though evolution were a tape recording that could be rerun over and over again to produce the same outcome. Kacar and Gaucher expected a similar outcome in this case. “Since the ancient gene is a direct ancestor, I naively thought it would simply accumulate mutations that make it look like the modern E. coli gene sequence,” Gaucher said.
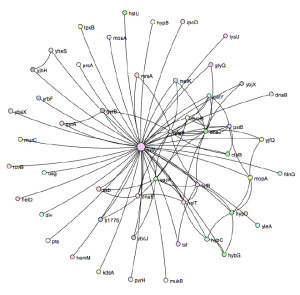
Kacar and Gaucher, Artificial Life (2012)
The protein elongation factor Tu, which helps to make new proteins, interacts with more than 50 other proteins in the cell, as shown here.
They were mistaken. In all eight of the E. coli strains they studied, the EF-Tu protein remained unchanged. Instead, seven strains changed the way the protein was used. Specifically, the E. coli developed genetic changes that made them pump out more of the subpar protein. “Making more of the ancient protein allows the cell to re-establish protein-protein interactions that had been disrupted,” Gaucher said.
In the eighth strain, some of the proteins that interact with EF-Tu changed so that they could better bind to the ancient version. It was as if the software in a modern laptop adapted to work better with an outdated processor. “The organisms that survive and pass on their genes to the next generation seem to be coping with the insertion of the ancient version not by reverting back to the modern version but by changing the other proteins that interact with it,” said Aaron Goldman, a computational biologist at Oberlin College in Ohio, who wasn’t involved in the study.
The findings inform an ongoing debate in evolution: Does adaptation proceed through changes to the parts of genes that make proteins, or through changes to the genome’s so-called regulatory regions, which control when, where and how much protein is made? In short, does evolution rely most on altering the proteins themselves, or does it depend on changing the way in which the cell uses proteins?
As is often the case in the complicated world of biology, Kacar’s results suggest that the answer is both. But regulatory mutations likely provide an easier route for improving fitness. Boosting protein production is easier than creating a more efficient version of the protein itself.
Network Hub
Kacar’s experiment appears to be the first in which an ancient version of a protein was successfully inserted into a living organism that was then allowed to evolve. That’s an extremely complicated endeavor, much more difficult than getting a protein to function in a test tube. The fact that the ancient protein works at all in a modern environment is itself significant, said Greg Fournier, a geobiologist at the Massachusetts Institute of Technology who was not involved in the research.
Related Article:
How Structure Arose in the Primordial Soup
Life’s first epoch saw incredible advances — cells, metabolism and DNA, to name a few. Researchers are resurrecting ancient proteins to illuminate the biological dark ages.
But that environment allows scientists to study the evolution of the whole complex network that surrounds the EF-Tu hub. “When we think about evolution, we tend to think about it at the level of the organism, or the genes,” Goldman said. “But they have captured a result that speaks to the in-between level.” This can offer insight into how molecular context — the various components of E. coli’s cellular machinery — affects evolution.
As a hub protein — more than 50 other proteins bind to it — EF-Tu appears to be resistant to change, forcing its molecular partners to adapt instead. This suggests that the position of a protein within a network determines whether it’s likely to be conserved or malleable over time, Goldman said. “To my knowledge, this is the first really clear view of that process.”
The unique eighth strain of E. coli — the one in which other proteins evolved — has already given researchers insight into the EF-Tu protein’s role in the cell. By examining the proteins that adapted to the ancient version, Kacar and collaborators were able to identify new binding partners for EF-Tu. “What is especially fascinating about Betul’s work is how the mutant screen identified new interacting partners,” Fournier said.
It appears that these partner proteins may have taken a step back in time to match the ancient transplant, raising hopes that the researchers might be able to piece together the ancient molecular network. “You can’t just resurrect an ancient gene and understand it,” Kacar said. “You need to resurrect the ancient metabolic pathway.”
Kacar next plans to apply the same approach much more broadly. With collaborators at Uppsala University in Sweden, she plans to replace modern E. coli genes with more ancient versions, but also to insert foreign versions from other species. She’ll look for evolutionary patterns as she travels back in time or further from E. coli on the evolutionary tree. These are her fragments of evidence, her relics of the distant past: alive and malleable, able to reveal the story of life.

![[No Caption]](https://www.quantamagazine.org/wp-content/uploads/2015/04/ChemicalAlphabetSoup-300x195.jpg)