Skip to comments.
It's Fun Seeing Evolution Falsified
CreationEvolutionHeadlines ^
| October 8, 2008
Posted on 10/08/2008 7:21:40 AM PDT by GodGunsGuts
click here to read article
Navigation: use the links below to view more comments.
first previous 1-20 ... 161-180, 181-200, 201-220 ... 321-324 next last
To: AndrewC
Many genes of known function and under high selection can be removed in a “knock out” mouse to no ill effect. For just one of hundreds of examples, a protein thought by a lab to work in the heart stress response was removed and the mouse looked just fine. It took severe stress to sort of “tease out” the phenotype, and even then it was pretty weak. No doubt these “knock out mice” will be found to have a difference as well.
Also if your read the original ScienceNews article these conserved sequences are believed to act as an enhancer region (promoting transcription) of a critical gene.
So are you betting the farm that no function will be found for these conserved regions? Or that the function won't be “critical enough” to satisfy you as an explanation for their similarity between species?
What explanation do you have for the presence of conserved, ultraconserved, and nonconserved regions in the genome?
Why would they follow with mathematical precision a divergence from commonality when comparing conserved to conserved, nonconserved to nonconserved, etc, exactly as if species shared a common ancestor with both types of sequences changing at divergent rates?
Seriously, if you want to break the link between conservation between species and function it is nearly impossible at this point to do it with negative (so far)evidence. You need POSITIVE evidence breaking the linkage.
So go out and find a nonconserved sequence that has a function. Should be easy if you agree with the Creationsafari blog that all DNA is functional. Heck, it that was true then almost any DNA will do.
Until you actually present positive evidence of your claim of the linkage being broken, you can either staking a claim that no function will ever be found for this particular DNA sequence, or that the function isn't ‘critical’ enough to explain the extent of its conservation between species.
But entire genes of known function and use, and with evolutionary conservation between species (let alone enhancers of a critical gene) have been shown to be removable and still produce a healthy happy mouse.
181
posted on
10/13/2008 1:01:29 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream; AndrewC
==The problem with your stupid dolls is that the data from one doesn't fit into the other no matter how hard you would try.
As for the Russian nested dolls, they were merely my first, and simplest example of intelligent designers deliberately creating nested hierarchies.
I see you completely avoided the questions I asked with respect to the nested hierarchy of the much more sophisticated military organization chart I posted. Why is that, Allmendream?
=Yes, I think miRNA (not unique to humans, do you still not understand what “highly conserved between species” means, should I explain it to you again?) could and did evolve over several million years.
I was specifically referring to the 51 miRNA that ARE UNIQUE TO HUMANS! miRNA are 21 to 23 nucleotides in length. According to Evo calculations, base pair mutation rates are between 10-8 and 10-10 PER GENERATION! So tell me Allmendream, how many millions upon millions of generations would it take for humans to develop 51 unique miRNA not shared by chimps? And that's not even counting the hundreds of cognate mutations that would have to occur at precise locations with respect to the hundreds of mRNA they regulate. Needless to say, this does not square with the Darwinian notion that humans diverged from chimps around 6 million years ago, now does it!
Like I said, you guys will believe ANYTHING so long as it papers-over God's handiwork.
To: GodGunsGuts
You are still so ignorant of the subject that you believe anyone who accepts the evidence for evolution wants to deny God? Like me and the Pope and the majority of Christians and the thousands of Scientist who are Christian, perhaps?. Your bias is clear. You cannot force yourself to accept the data because to you that would be denying God. Your religious objections are noted and they do nothing but subtract from your credibility.
The miRNA data you sourced says the sequences are HIGHLY CONSERVED BETWEEN SPECIES. It doesn't say that they are unique to humans. Perhaps you mean the 41 miRNA’s (not 51) found in lampreys and subsequently also found in mice (in case your still not following that would mean that they are HIGHLY CONSERVED BETWEEN SPECIES GGG), is that what you meant?
Still not addressing the question. There is no explanation other than common ancestry for these nested hierarchies, or for the Vultures not being similar to each other. You have not even attempted an explanation that holds up or even addresses the issue.
And the data on genome homology supports the figure of six million years for our divergence from chimps.
183
posted on
10/13/2008 2:34:49 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: GodGunsGuts
Wow. Once again conservation between species and function is shown to be a powerfully predictive tool in regards to miRNA and their role in translation regulation. Is anyone surprised at any FURTHER confirmation of the linkage between conservation between species and function?
Human MicroRNA targets.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS.
Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
MicroRNAs (miRNAs) interact with target mRNAs at specific sites to induce cleavage of the message or inhibit translation. The specific function of most mammalian miRNAs is unknown. We have predicted target sites on the 3’ untranslated regions of human gene transcripts for all currently known 218 mammalian miRNAs to facilitate focused experiments. We report about 2,000 human genes with miRNA target sites conserved in mammals and about 250 human genes conserved as targets between mammals and fish. The prediction algorithm optimizes sequence complementarity using position-specific rules and relies on strict requirements of interspecies conservation. Experimental support for the validity of the method comes from known targets and from strong enrichment of predicted targets in mRNAs associated with the fragile X mental retardation protein in mammals. This is consistent with the hypothesis that miRNAs act as sequence-specific adaptors in the interaction of ribonuclear particles with translationally regulated messages. Overrepresented groups of targets include mRNAs coding for transcription factors, components of the miRNA machinery, and other proteins involved in translational regulation, as well as components of the ubiquitin machinery, representing novel feedback loops in gene regulation. Detailed information about target genes, target processes, and open-source software for target prediction (miRanda) is available at http://www.microrna.org. Our analysis suggests that miRNA genes, which are about 1% of all human genes, regulate protein production for 10% or more of all human genes.
PMID: 15502875 [PubMed - indexed for MEDLINE]
PMCID: PMC521178
184
posted on
10/13/2008 2:46:07 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream; AndrewC
==So go out and find a nonconserved sequence that has a function. Should be easy if you agree with the Creationsafari blog that all DNA is functional.
We also found that three of five unconserved Ndt80 binding sites show Ndt80-dependent effects on gene expression. Together these data imply that although sequence conservation can be reliably used to predict functional TFBSs, unconserved sequences might also make a significant contribution to a species’ biology.
http://biblioteca.universia.net/ficha.do?id=5080356
This trans-activating activity is localized to a 35 amino acid segment within the evolutionarily unconserved central region.
http://www.springerlink.com/content/p3220113v6223023/
These results show that the two isolated Ig- and Ig- ITAMs are able to trigger the same intracellular events leading to cytokine production and that the unconserved environment of an ITAM can regulate the signaling activity of this kind of tyrosine-based activating motifs.
http://www.jbc.org/cgi/content/full/271/39/23786
To: allmendream; AndrewC
==You are still so ignorant of the subject that you believe anyone who accepts the evidence for evolution wants to deny God? Like me and the Pope and the majority of Christians and the thousands of Scientist who are Christian, perhaps?
Speaking of Christians, you never did say what church you attend since leaving the Armenian Orthodox Church. Do you still belong to a church?
==Your bias is clear. You cannot force yourself to accept the data because to you that would be denying God.
Interesting, I was thinking the exact same thing about you. You cannot force yourself to accept the Biblical creatrion account, or the evidence that back up the same, because it force you to deny evolution.
==The miRNA data you sourced says the sequences are HIGHLY CONSERVED BETWEEN SPECIES. It doesn't say that they are unique to humans. Perhaps you mean the 41 miRNA’s (not 51) found in lampreys and subsequently also found in mice (in case your still not following that would mean that they are HIGHLY CONSERVED BETWEEN SPECIES GGG), is that what you meant?
Nope, I meant what I said. Fifty-one miRNA sequences found in humans are not found in chimps. From an Evo point of view, this means that 51 uniquely human miRNAs, and a network of hundreds of additional cognate mutations for each miRNA (at a base pair mutation rate of 10-8and 10-10 per generation!), would have had to evolve in the last 6 million years. Is this what you believe, Allmendream?
For more, see the following:
http://www.ncbi.nlm.nih.gov/pubmed/17072315
Finally, why do you insist that only conserved sequences will have function??? If only conserved sequences had function, wouldn't that reduce the "tree of life" to a single specie?
To: GodGunsGuts
Sequence conservation can be reliable used to predict functional Transcription Factor Binding Domains? Wow. Once again the link holds up.
The central region of that protein has a transacting domain. The central region of 35 amino acids is relatively unconserved compared to the rest of the protein.
“We showed that BCR subunits transducing activities are based on the ITAM conserved sequence, but they may be regulated by unconserved sequences located inside or outside the motif.”
The activity is based on conserved sequences? Wow. Once again. May be regulated by unconserved sequences.
187
posted on
10/13/2008 3:49:56 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: GodGunsGuts
Where are these 51 unique to human miRNA’s mentioned? Seems they found them in humans AND chimp brains, and said their conservation between species didn't extend past primates, giving them a recent origin, and that some were species specific. So out of the 447 found in common between human and chimps, your saying there are 51 unique to humans? This pattern would be quite consistent with their NARROW conservation between species; only among primates.
And no. Functional domains are conserved between related species; that doesn't mean they are exactly the same over the entire tree of life. Once again you don't even understand the subject. It is the PATTERN of SIMILARITY and DIFFERENCE which grows at a consistent rate depending upon the type of DNA as the relatedness of the species increases in the scope of comparison.
1: Nat Genet. 2006 Dec;38(12):1375-7. Epub 2006 Oct 29. Links
Diversity of microRNAs in human and chimpanzee brain.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH.
Hubrecht Laboratory-KNAW, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands.
We used massively parallel sequencing to compare the microRNA (miRNA) content of human and chimpanzee brains, and we identified 447 new miRNA genes. Many of the new miRNAs are not conserved beyond primates, indicating their recent origin, and some miRNAs seem species specific, whereas others are expanded in one species through duplication events. These data suggest that evolution of miRNAs is an ongoing process and that along with ancient, highly conserved miRNAs, there are a number of emerging miRNAs.
PMID: 17072315 [PubMed - indexed for MEDLINE]
188
posted on
10/13/2008 4:00:19 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream
==Sequence conservation can be reliable used to predict functional Transcription Factor Binding Domains? Wow. Once again the link holds up.
Can you read, Allmendream? So what if sequence conservation is a reliable predictor of function? You said “go out and find a nonconserved sequence that has a function.” So I went out and found nonconserved sequences that have function:
These results show that the two isolated Ig- and Ig- ITAMs are able to trigger the same intracellular events leading to cytokine production and that the unconserved environment of an ITAM can regulate the signaling activity of this kind of tyrosine-based activating motifs.
http://www.jbc.org/cgi/content/full/271/39/23786
The point being made is that non-conserved sequences are increasingly being shown to have function as well. Needless to say, Creation Science predicts functional sequences that are “unconserved” between species, whereas this discovery flies in the face of Darwinist predictions.
To: GodGunsGuts
Every gene in a human has a homologous gene in a chimp, none of them are unique. There are regulatory sequences that are unique, thus nonconserved between species. The regulatory function of none of these nonconserved sequences is essential to life, just essential to making us distinctly human; and the majority of nonconserved sequences show no function at all.
The challenge that would break the link between function and conservation between species was in a Knock out mouse, (you might remember the subject of this thread perhaps?); knock out a sequence unique to a mouse and see if it produces a phenotype. That would be positive evidence of an essential function in a nonconserved sequence, and would help break the overwhelming linkage Biologists see in THOUSANDS of experiments between function and evolutionary conservation between species.
190
posted on
10/13/2008 4:36:10 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream; GodGunsGuts
So are you betting the farm that no function will be found for these conserved regions? No, I've stated over and over that I believe that they have function.
What is at the heart of the matter is why the ultra-conservation. Darwinian evolution does not have an answer and it asserts that it does have an answer.
The purity of the regions from species to species speaks volumes as to the repair and maintenance of those regions. Darwininian theory requires natural selection to "purify" those regions. The fact that natural selection has, at the moment, nothing to work on is evidence that the Darwinian explanation is wrong. Even a weak selection does not explain the purity of the regions from species to species. Normal mutation rates require some modification to the genome, again, since the Darwinian explanation requires natural selection to "purify" the regions.
What explanation do you have for the presence of conserved, ultraconserved, and nonconserved regions in the genome?
A computing cell.
So go out and find a nonconserved sequence that has a function.
That is too easy, any gene for a protein unique to a species.
But entire genes of known function and use, and with evolutionary conservation between species (let alone enhancers of a critical gene) have been shown to be removable and still produce a healthy happy mouse.
So what? The question is of degree of conservation and removability, not of removability by itself.
Until you actually present positive evidence of your claim of the linkage being broken, you can either staking a claim that no function will ever be found for this particular DNA sequence, or that the function isn't ‘critical’ enough to explain the extent of its conservation between species.
The linkage is broken. The ultra-conserved regions were removed with no noticeable harm to the organisms. The paper that I cited previously depends entirely on the tying of degree of conservation to criticality. They bet on it.
This should allow us to define the lowest level of sequence variation in the human genome.
191
posted on
10/13/2008 6:55:32 PM PDT
by
AndrewC
To: AndrewC
There is an answer and it is an easy one. These regulatory sequences, like many genes that can also be removed without a noticeable phenotype, are under selection that is not obvious in a lab raised mouse.
The linkage between conservation between species and function is one that has held true thousands of times. Conservation between species in genome comparison has been in invaluable aid in “homing in” on functional sequences.
If the issue is degree of conservation and “removeability” then why the “so what?” to the fact that entire genes can be removed. A gene is not “functional” enough for you?
Biologist aren't going to ignore a hundred thousand good examples of the linkage between function and conservation between species just because one highly conserved regulatory sequence has, so far, not produced a phenotype when removed.
You think cells are self aware and can direct their own evolution? Wow, can you pass that stuff this way?
192
posted on
10/13/2008 7:40:58 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream; AndrewC
==Where are these 51 unique to human miRNA’s mentioned? Seems they found them in humans AND chimp brains, and said their conservation between species didn't extend past primates
Did you even bother reading the paper? Look, here's a blowup of the charts in question. Take a look at ITEM C. It clearly demonstrates that there are 51 MicroRNAs that are unique to humans, and 25 MicroRNAs that are unique to Chimpanzees. Next time I send you a link to a paper, you might want to read it before accusing me of not knowing what I'm talking about. Which brings me back to the the question that you conveniently declined to answer:
From an Evo point of view, this means that 51 uniquely human miRNAs, and a network of hundreds of additional cognate mutations for each miRNA (at a base pair mutation rate of 10-8 and 10-10 per generation!), would have had to evolve in the last 6 million years. Is this what you actually believe, Allmendream?
Again, here is the chart, PAY CLOSE ATTENTION TO ITEM C.
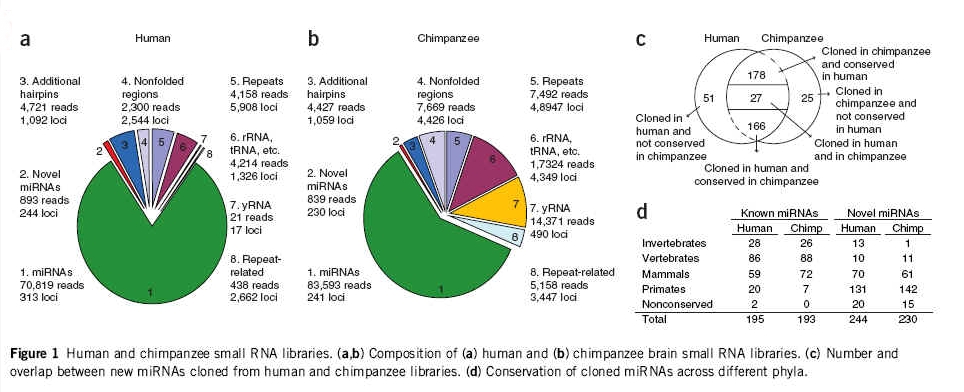
To: GodGunsGuts
No I didn't see that in the abstract you linked. Can you provide the link to the entire paper?
Yes, I believe these regulatory sequences evolved over six million years. Do you think a 2% genetic change and a 6% genomic change is impossible to accomplish in six million years?
Oh yeah, you don't even believe the world existed six million years ago.
194
posted on
10/13/2008 8:44:36 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: AndrewC
==No, I've stated over and over that I believe that they have function.
He keeps trying to straight-jacket me with false choices too. Apparently Allmendream is having a hard time coming to terms with the fact that a sequence can be "ultraconserved" between species, and yet not be critical to the life of the organism. He just can't grasp the idea that the sequence could just as well be specified by the organism's biotic program.
To: GodGunsGuts
These guys think the sequences evolved over six million years or so as well.
http://genome.cshlp.org/cgi/content/full/17/5/612?ck=nck
Rapid evolution of an X-linked microRNA cluster in primates
Rui Zhang1,2,3, Yi Peng1,2, Wen Wang1, and Bing Su1,2,4
MicroRNAs (miRNAs) are a growing class of small RNAs (about 22 nt) that play crucial regulatory roles in the genome by targeting mRNAs for cleavage or translational repression. Most of the identified miRNAs are highly conserved among species, indicating strong functional constraint on miRNA evolution. However, nonconserved miRNAs may contribute to functional novelties during evolution. Recently, an X-linked miRNA cluster was reported with multiple copies in primates but not in rodents or dog. Here we sequenced and compared this miRNA cluster in major primate lineages including human, great ape, lesser ape, Old World monkey, and New World monkey. Our data indicate rapid evolution of this cluster in primates including frequent tandem duplications and nucleotide substitutions. In addition, lineage-specific substitutions were observed in human and chimpanzee, leading to the emergence of potential novel mature miRNAs. The expression analysis in rhesus monkeys revealed a strong correlation between miRNA expression changes and male sexual maturation, suggesting regulatory roles of this miRNA cluster in testis development and spermatogenesis. We propose that, like protein-coding genes, miRNA genes involved in male reproduction are subject to rapid adaptive changes that may contribute to functional novelties during evolution.
196
posted on
10/13/2008 9:00:43 PM PDT
by
allmendream
(White Dog Democrat: A Democrat who will not vote for 0bama because he's black.)
To: allmendream
To: allmendream
==These guys think the sequences evolved over six million years or so as well.
I’ll say! You guys believe in materialist miracles!!! From your paper:
“Our data indicate rapid evolution of this cluster in primates including frequent tandem duplications and nucleotide substitutions.”
To: allmendream; AndrewC
==You think cells are self aware and can direct their own evolution?
He didn't say self aware. He specifically made reference to the notion of a computing cell. Are computers self-aware?
To: allmendream
PS Why are you afraid to say which church you belong to now? Have you stopped going to church? Joined a cult? Why the evasiveness?
Navigation: use the links below to view more comments.
first previous 1-20 ... 161-180, 181-200, 201-220 ... 321-324 next last
Disclaimer:
Opinions posted on Free Republic are those of the individual
posters and do not necessarily represent the opinion of Free Republic or its
management. All materials posted herein are protected by copyright law and the
exemption for fair use of copyrighted works.
FreeRepublic.com is powered by software copyright 2000-2008 John Robinson
